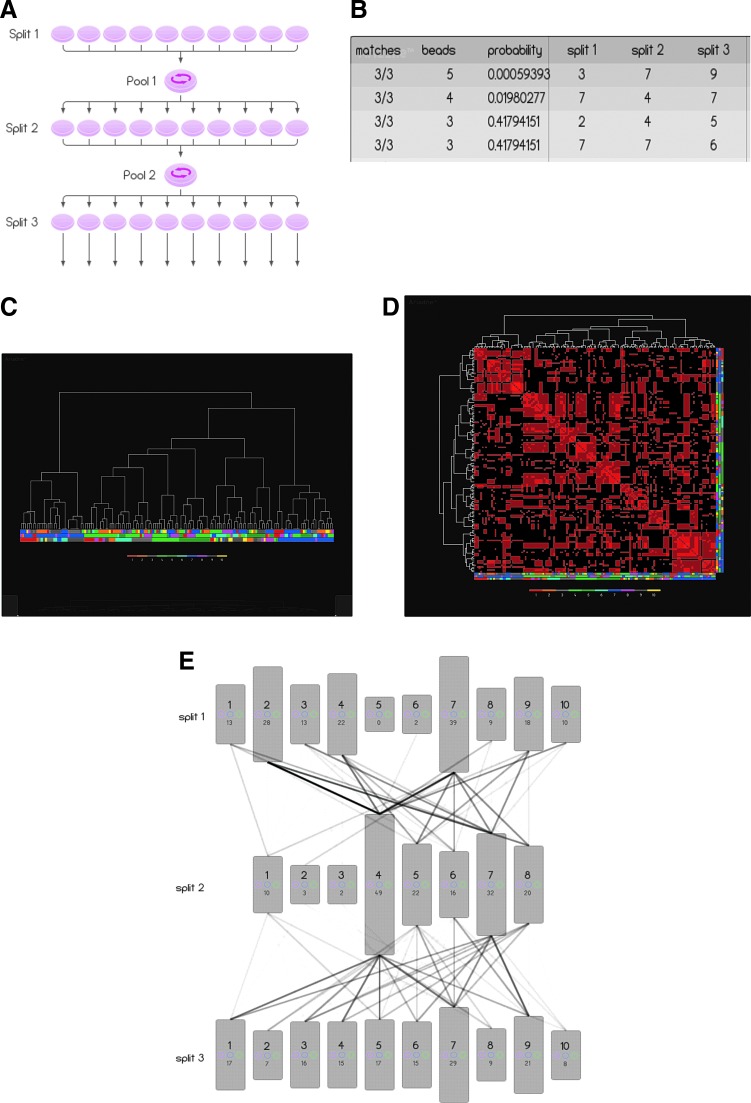
FIG. 2.
CombiCult screen and Ariadne analysis. (A) CombiCult matrix scheme showing the split-pool schedule and number of conditions sampled. Ariadne analysis of the CombiCult screen showing (B) the fingerprint analysis: each row of the table represents a group of hits clustered according to protocol similarity. Column 1 identifies the number of culture conditions the group has in common; column 2, the number of hits included in the group; and column 3 the probability of the group occurring by chance. Columns 4–6 identify common media in the groups. (C) Hierarchical clustering analysis of 124 unique protocols derived from the screen, showing all protocol clusters. Each node at the bottom of the dendrogram (leaf node) corresponds to a hit bead. The associated protocol is denoted by the column of four colors directly below the node, specifying the media sampled in splits 1–3. The legend at the bottom of the figure specifies the color used to denote the cell culture media in each split. (D) The similarity matrix comprising a pairwise comparison of all protocols. Each column and each row corresponds to a protocol. The brightness of each cell in the matrix is proportional to the number of identical cell culture media shared by the two protocols. The brightest cell corresponds to identical protocols, while a black cell corresponds to two protocols with no common media. The diagonal row of cells (from the top left to bottom right) corresponds to protocols being compared with themselves. Protocol families with high internal homology appear as bright red squares. (E) Schematic diagram illustrating an overlay of all protocols deconvoluted from 154 hits. The height of boxes representing each cell culture medium is proportional to the number of hits generated by that medium (written at the bottom of each box). The opacity of the linkage lines is proportional to the number of hits generated by specific medium combinations—the darkest line corresponds to 12 hits.