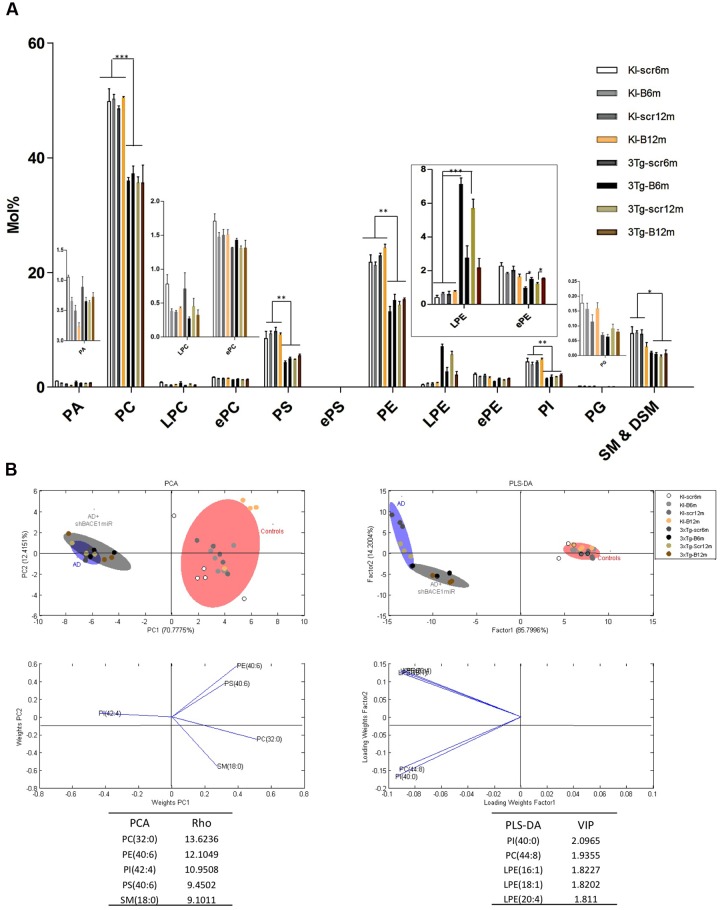
FIGURE 3.
Lipid composition of the hippocampus from 3xTg-AD and control mice, grouped according to the time of treatment. Various lipid changes can be seen in the hippocampal tissue. (A) The lipid class profiles are expressed as %mol composition. All lipid species were measured (means), and the error bars represent the SEM. The data for the 3xTg-AD mice were significantly different from the control groups (∗p < 0.05, ∗∗p < 0.01; ∗∗∗p < 0.001 ANOVA followed by the Tukey post hoc test or Kruskal–Wallis test). (B) Multivariate analyses of the lipid profiles from the hippocampus. PCA, principal component analyses for the lipid classes; PLS-DA, partial least squares analysis to discriminate between the lipid classes. The left panels illustrate the factor loadings for PC1 and PC2, with the indices of variance explained for each component. The right panels show the factor score plots for PLS-DA. The variables in the analyses are the controls, AD (3xTg-AD without treatment) and AD+shBACE1-miR (3xTg-AD with 6 and 12 months of treatment). PA, phosphatidic acid; PC, phosphatidylcholine; LPC, lysophosphatidylcholine; ePC etherphosphatidylcholine; PS, phosphatidylserine; ePS, etherphosphatidylserine; PE, phosphatidylethanolamine; LPE, lysophosphatidylethanolamine; ePE, etherphosphatidylethanolamine, PI, phosphatidylinositol; PG, phosphatidylglycerol; SM, sphingomyelin. n = 3–5 per group.