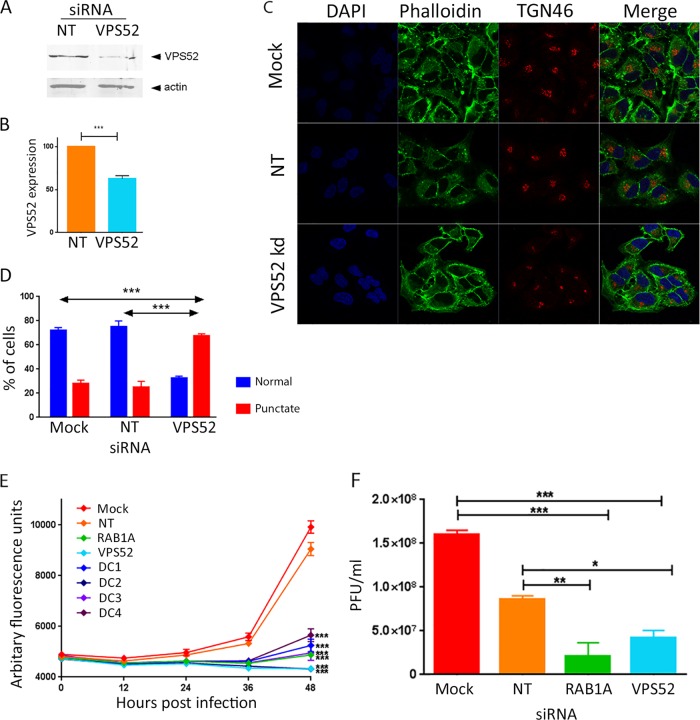
FIG 1.
Reduced expression of VPS52 impairs multistep growth of VACV. (A) HeLa cells were transfected with nontargeting siRNA (NT) or a SMARTpool of four siRNAs targeting VPS52. After 48 h, cells were harvested and proteins were separated using SDS-PAGE, probed with antibody raised against VPS52 or actin, and visualized using direct infrared fluorescence (LI-COR) in an Odyssey scanner. (B) The levels of VPS52 relative to those of actin were quantified. Data represent the averages from three biological replicates. Error bars are SEMs. ***, P < 0.001 by two-tailed Student's t test. (C) HeLa cells were mock transfected or transfected with nontargeting siRNA or siRNA targeting VPS52. After 48 h, cells were fixed and labeled with antibody raised against TGN46 and phalloidin. (D) The percentage of cells with a normal punctate TGN46 distribution was calculated. ***, P < 0.001 by two-tailed Student's t test. (E) HeLa cells were transfected with a VPS52 siRNA SMARTpool (VPS52) and the four individual deconvoluted VPS52 siRNAs (DC1 to DC4). Negative controls included mock-transfected cells and cells transfected with nontargeting siRNA. siRNA targeting a known proviral host factor (RAB1A) was used as the positive control. After 48 h, cells were infected with VACV-EGFP at 0.1 PFU/cell, and fluorescence levels were measured over the following 48 h. The data represent those from six technical replicates and are representative of those from three biological replicates. Error bars represent SEMs. Results were analyzed with a one-way analysis of variance with multiple comparisons at 48 h postinfection. ***, P < 0.001. (F) HeLa cells were mock transfected or transfected with nontargeting siRNA or siRNA targeting VPS52 or RAB1A. After 48 h, cells were infected with VACV-EGFP at 0.1 PFU/cell, and at 48 h p.i., cells and supernatant were collected and virus titers were determined. Data were analyzed with a one-way analysis of variance with multiple comparisons. ***, P < 0.001; **, P < 0.01; *, P < 0.05.