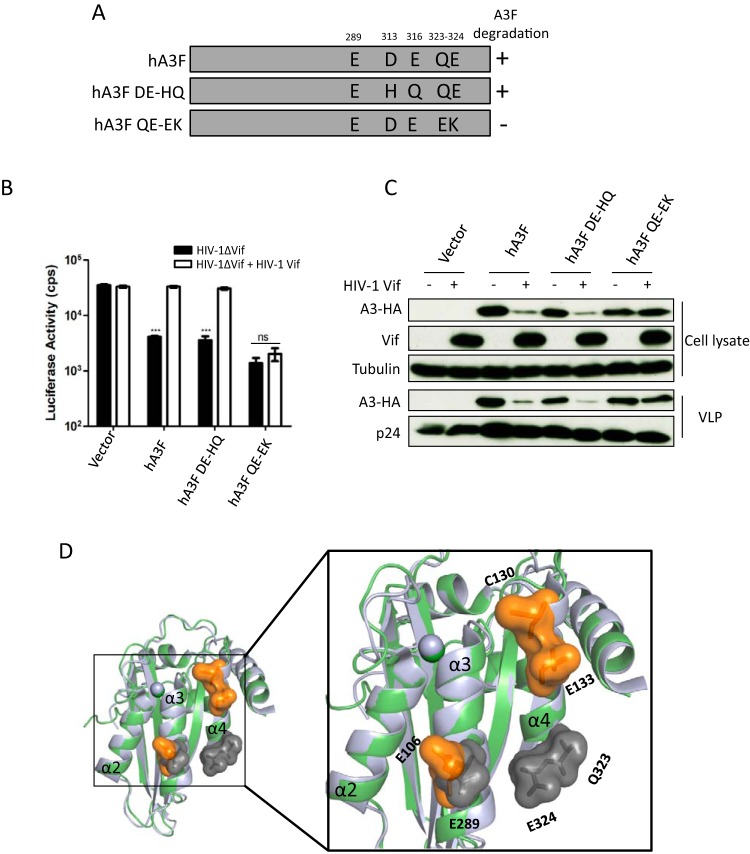
FIG 5.
hA3F residues and HIV-1 Vif-induced depletion. (A) The schematic structure of hA3F mutants. +, sensitive to HIV-1 Vif induced degradation; −, resistant to HIV-1 Vif-induced degradation. (B) HIV-1ΔVif luciferase or HIV-1ΔVif luciferase plus HIV-1 Vif were produced in 293T cells in the presence of A3F or mutants. After normalizing for reverse transcriptase activity, viral infectivity was determined by quantification of luciferase activity in 293T cells. (C) A3s in HIV-1 producer cells and HIV-1 viral particles were detected by using anti-HA antibody. HIV-1 Vif and capsid (p24) were detected by anti-Vif and anti-p24 antibodies, respectively. Tubulin served as a loading control. (D) Superimposition of A3C crystal structure (PDB entry 3VOW) (green) and A3F-CTD crystal structure (PDB entry 4J4J) (light blue). Key residues E106, C130, and E133 are shown as orange surfaces in A3C with labels, and the essential residues (E289, Q323, and E324) in A3F are highlighted with gray surfaces and labels. cps, counts per second. Asterisks represent statistically significant differences: ***, P < 0.001; ns, no significance (Dunnett's t test).