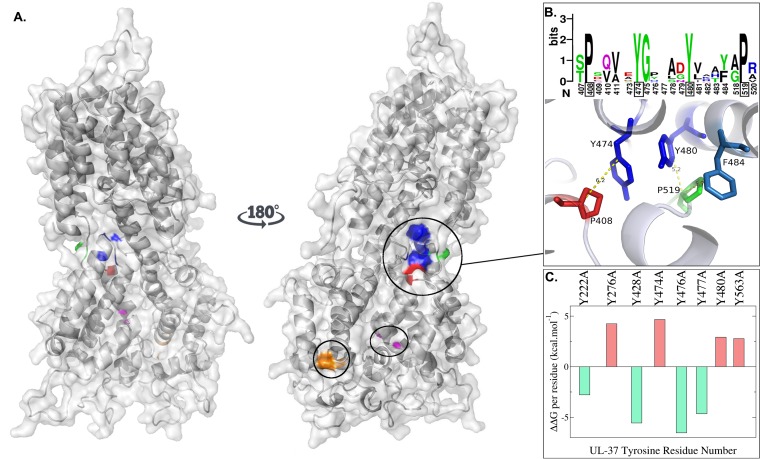
FIG 3.
Predicted structure and dynamics of the HSV-1 UL37 protein. (A) Space-filling predictive model of 570 amino acids in the amino terminus of pUL37 in HSV-1 based on the known crystal structure of a corresponding amino-terminal region of PRV UL37. Blue, Y476-Y477 and Y480; orange, P262-L263; red, P408; green, P519; magenta, G419-F420. (B) Sequence logo depicts the relative conservation of amino acids 407 to 411, 473 to 484, and 518 to 520 (top portion of panel B). Approximate distances between the aromatic residues Y474, Y480, and F484 and P408 and P519 (bottom portion of panel B). (C) In silico alanine-scanning mutagenesis. The difference between the free Gibbs energy before and after the mutation (ΔΔG) is shown on the y axis. Values are reported as the deviation from the median ΔΔG of UL37 tyrosine-to-alanine mutations. Negative values indicate stabilizing mutations (green). Positive values indicate destabilizing mutations (red).