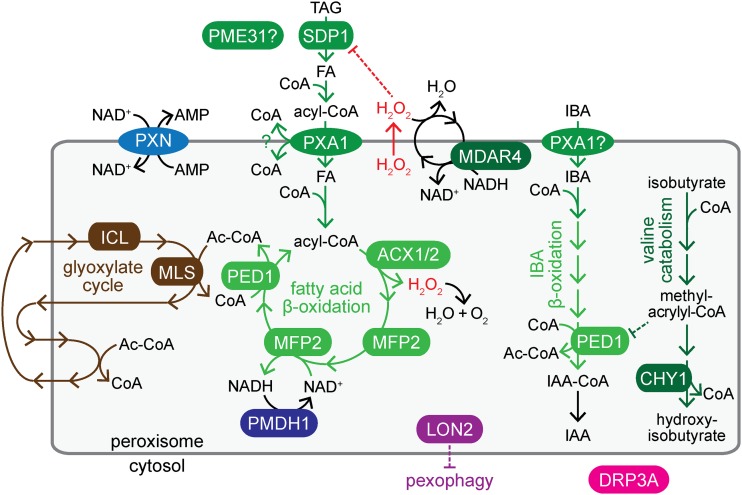
Figure 1.
Peroxisomes house steps of key metabolic pathways; enzymes and transporters disrupted in mutants identified in this work are shown. Conversion of oil body triacylglycerol (TAG) to fatty acids (FA) requires SDP1. Fatty acids are activated with CoA in the cytosol and imported into peroxisomes by PXA1, which hydrolyzes the CoA upon transport. In the peroxisome, fatty acids are reactivated and catabolized to acetyl-CoA through β-oxidation (green), which requires ACX1/2, MFP2, and PED1 enzymes. ACX enzymes generate H2O2, which is detoxified by catalase (not shown), or by an MDAR4-dependent ascorbate peroxidase that inactivates H2O2 escaping from the peroxisome. CHY1 acts in valine catabolism downstream of methacrylyl-CoA, which can inactivate PED1. PME31 might aid in lipid mobilization. PXA1 is also implicated in peroxisomal import of IBA, and IBA-to-IAA β-oxidation likely involves dedicated enzymes (not shown) and PED1, which also acts in fatty acid β-oxidation. Carbon from β-oxidation-derived acetyl-CoA is converted to forms that can be shuttled out of the peroxisome via the glyoxylate cycle (brown), which includes ICL and MLS enzymes. PXN imports NAD+, which is recycled by PMDH1/2; CoA is recycled in the glyoxylate cycle. LON2 is an ATP-dependent protease that restrains pexophagy. DRP3A is a peroxisome division factor.