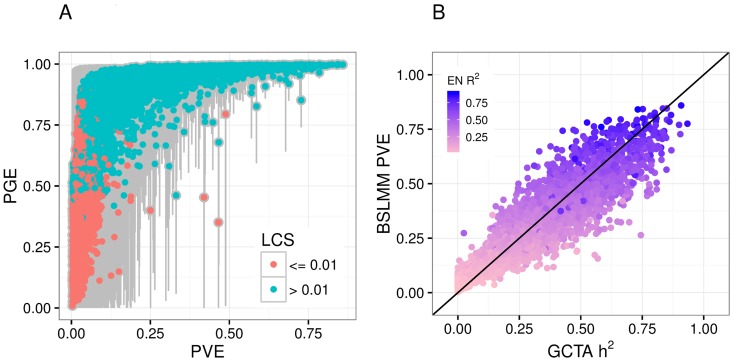
Fig 2. Sparsity estimates using Bayesian Sparse Linear Mixed Models in DGN whole blood.
(A) This panel shows a measure of sparsity of the gene expression traits represented by the PGE parameter from the BSLMM approach. PGE is the proportion of the sparse component of the total variance explained by genetic variants, PVE (the BSLMM equivalent of h2). The median of the posterior samples of BSLMM output is used as estimates of these parameters. Genes with a lower credible set (LCS) > 0.01 are shown in blue and the rest in red. The 95% credible set of each estimate is shown in gray. For highly heritable genes the sparse component is close to 1, thus for high heritability genes the local architecture is sparse. For lower heritability genes, there is not enough evidence to determine sparsity or polygenicity. (B) This panel shows the heritability estimate from BSLMM (PVE) vs the estimates from GCTA, which are found to be similar (R = 0.96). Here, the estimates are constrained to be between 0 and 1 in both models. Each point is colored according to that gene’s elastic net α = 1 cross-validated prediction correlation squared (EN R2). Note genes with high heritability have high prediction R2, as expected.