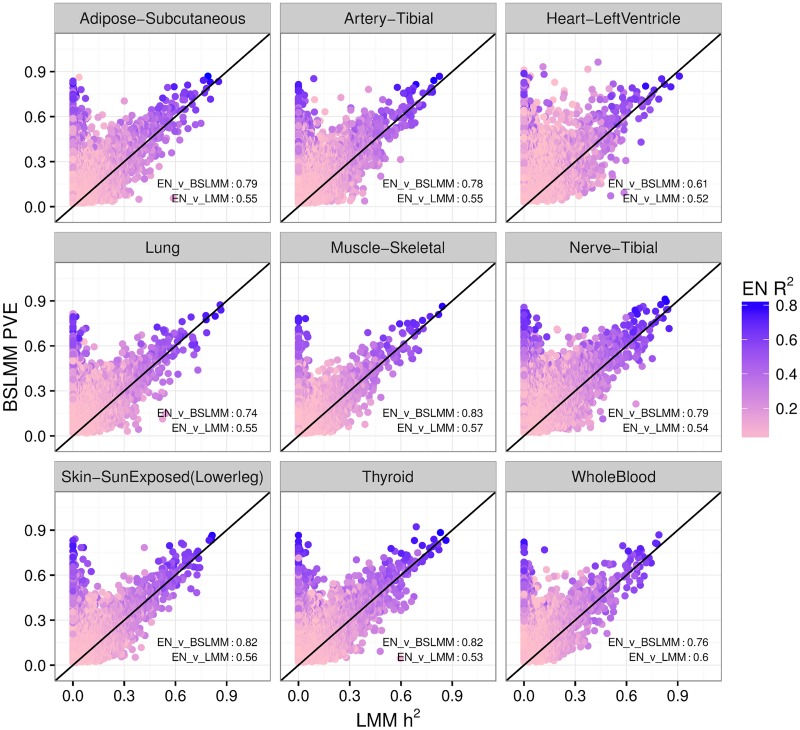
Fig 4. BSLMM vs LMM estimates of heritability in GTEx.
This figure shows the comparison between estimates of heritability using BSLMM vs. LMM (GCTA) for GTEx data. Here, in both models the estimates are constrained to be between 0 and 1. For most genes BSLMM estimates are larger than LMM estimates reflecting the fact that BSLMM yields better estimates of heritability because of its ability to account for the sparse component. Each point is colored according to that gene’s prediction R2 (correlation squared between cross-validated elastic net prediction vs observed expression denoted EN R2). At the bottom right of each panel, we show the correlation between BSLMM (EN_v_BSLMM) and LMM (EN_v_LMM). BSLMM is consistently more correlated with the elastic net correlation. This provides further indication that the local architecture is predominantly sparse.