Fig. 1. Nucleosomes cause strong pausing of the T7 replisome.
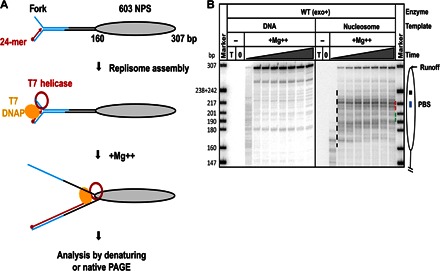
(A) Experimental approach for analysis of replication through chromatin. Each template contains the fork DNA structure, linker DNA, and the strong 603 NPS. The fork DNA structure (in blue) is ligated to the nucleosomal template (gray oval). After ligation, the P32-end–labeled 24-mer DNA primer (red arrow) is annealed to the template DNA strand. Next, DNAP, which was preassembled with E. coli trx and T7 helicase (red ring), is added to the reaction and forms a replisome in the presence of all dNTPs. The reaction is then activated by the addition of Mg++. Replication products are analyzed by denaturing or native PAGE. (B) Analysis of labeled products after replication of 603 DNA (histone-free or organized in a nucleosome) for different time intervals (0, 2, 5, 10, 30, 60, 120, 240, and 480 s) by denaturing PAGE. The locations of the nucleosome (oval), the nucleosome dyad (square), the PBS (blue line), and the runoff transcript and nucleosome-specific pausing (black dashed line) are shown. T, DNA template only; 0, reaction before addition of Mg++. Note that the nucleosomal pausing regions +(31–40) and +(41–65) are indicated by green and red dashed lines, respectively. Markers are pBR322 DNA–Msp I digest marker (New England Biolabs). The sizes of marker DNA fragments are indicated on the left side.
