Abstract
Auxin directs plant ontogenesis via differential accumulation within tissues depending largely on the activity of PIN proteins that mediate auxin efflux from cells and its directional cell-to-cell transport. Regardless of the developmental importance of PINs, the structure of these transporters is poorly characterized. Here, we present experimental data concerning protein topology of plasma membrane-localized PINs. Utilizing approaches based on pH-dependent quenching of fluorescent reporters combined with immunolocalization techniques, we mapped the membrane topology of PINs and further cross-validated our results using available topology modeling software. We delineated the topology of PIN1 with two transmembrane (TM) bundles of five α-helices linked by a large intracellular loop and a C-terminus positioned outside the cytoplasm. Using constraints derived from our experimental data, we also provide an updated position of helical regions generating a verisimilitude model of PIN1. Since the canonical long PINs show a high degree of conservation in TM domains and auxin transport capacity has been demonstrated for Arabidopsis representatives of this group, this empirically enhanced topological model of PIN1 will be an important starting point for further studies on PIN structure–function relationships. In addition, we have established protocols that can be used to probe the topology of other plasma membrane proteins in plants.
Key words: plasma membrane protein, topology, auxin efflux carriers, Arabidopsis thaliana
Utilizing techniques based on pH-dependent quenching of fluorescent reporters and immunolocalizations we mapped PIN membrane topology, and further cross-validated our results with topology modeling software using empirical constraints. We delineated the topology of PIN1 with two transmembrane bundles of five α-helices linked by a large intracellular loop and an extracellular C-terminus.
Introduction
Auxin is an important regulator of plant development. Through its differential distribution within plant tissues, this signaling molecule provides instructive cues to guide plant development from embryogenesis (Vanneste and Friml, 2009, Adamowski and Friml, 2015, Robert et al., 2015) throughout the entire plant ontogenesis (Benková et al., 2003, Berleth et al., 2007, Benjamins and Scheres, 2008, Swarup et al., 2008). Local auxin distribution depends on auxin biosynthesis, conjugation, and degradation (Simon and Petrášek, 2011, Ljung, 2013), but also largely on auxin translocation between cells (Adamowski and Friml, 2015). A crucial aspect of this directional transport is mediated by the PIN auxin efflux components, whose family in Arabidopsis consists of PIN1, 2, 3, 4, and 7; these subclade members localize asymmetrically at the cell plasma membrane (PM) (Petrášek et al., 2006, Wiśniewska et al., 2006, Adamowski and Friml, 2015). The more distant and shorter subclades PIN5 and PIN8 are predominantly localized at the endoplasmic reticulum (ER) membrane, where they presumably mediate auxin exchange between the cytosol and the ER lumen (Mravec et al., 2009; Bosco et al., 2012, Ding et al., 2012) with possible minor additional localization at the PM in some cases (Ganguly et al., 2014). The localization and function of the intermediate member PIN6 is not yet entirely clarified (Nisar et al., 2014). The auxin transport capacity of PINs was demonstrated in both Arabidopsis and heterologous systems (Petrášek et al., 2006, Yang and Murphy, 2009, Zourelidou et al., 2014). These results are also supported by results of genetic interference with those proteins, leading to reduced auxin transport and developmental phenotypes similar to those observed after application of auxin transport inhibitors (Okada et al., 1991, Gälweiler et al., 1998, Vieten et al., 2007).
Given the importance of auxin transport in plant development and its directionality, a large body of experimental data concerns the clathrin-mediated endocytosis, subcellular trafficking including delivery to the vacuole, and polarization of PINs (Abas et al., 2006, Kleine-Vehn et al., 2008a, Kleine-Vehn et al., 2008b, Kitakura et al., 2011; for an overview see Adamowski and Friml, 2015). Comparably little is known about the structure of PINs. Their predicted amino acid sequences clearly indicate them as integral membrane proteins with several transmembrane (TM) domains. In addition, the topological models generated for these transporters all indicate a large hydrophilic region in the middle of the PM-localized PINs (Palme and Gälweiler, 1999). The loop (also often designated as hydrophilic loop and abbreviated to HL) has been shown to be phosphorylated by cytoplasmic kinases such as PINOID or D6PKs of the AGC kinase family (Friml et al., 2004, Michniewicz et al., 2007, Ganguly et al., 2014, Zourelidou et al., 2014). In contrast, the corresponding regions of ER-localized PINs harbor only a very short stretch of hydrophilic amino acids between the TM domains (Křeček et al., 2009, Mravec et al., 2009). Although there exists an assumed consensus about how PINs should look (Palme and Gälweiler, 1999, Ganguly et al., 2012, Bennett et al., 2014), there are surprisingly few experimental data to support any topology model.
Here, we experimentally tested the PIN1 topology while evaluating the likelihood of multiple software predictions. Our results show that the central loop of PIN1 localizes to the cytoplasm while its carboxy (C)-terminal end faces the apoplast, consistent with models indicating two regions with five TM domains spaced by a hydrophilic region. Using constraints derived from our empirical data, we also provide an updated position of the helical regions generating a verisimilitude model of PIN1.
Results
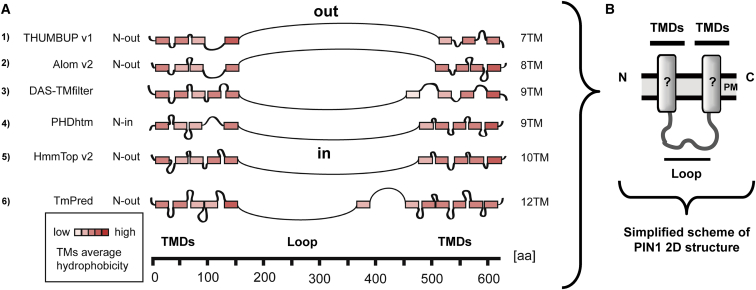
PIN1 Topology Predictions Vary in TM Domain Number but Indicate Consistently a Hydrophilic Region in the Central Part
While PIN protein localization and developmental functions are intensively studied, only limited information is available about the structure of these proteins. One reason for this status quo is the fact that PINs are TM proteins with multiple TM domains, and are therefore a demanding target for expression and purification procedures. This stimulated us to turn toward computational protein structure predictions. However, fold-recognition or template-based homology modeling (Schwede, 2013, Kelley et al., 2015) could not generate a reliable model for any of the PIN proteins (data not shown). Therefore, we resorted to more basic 2D topology predictive software, which, however, is known to be error-prone (Elofsson and von Heijne, 2007). This is especially true for ambiguities about the orientation of the N and C-termini (Jones, 2007). To grasp such variations, we generated a number of PIN1 topology predictions using different programs (Figure 1A) (Schwacke et al., 2003). The different simulations predicted PIN1 topologies having between 7 and 12 presumptive TM domains, resulting in variable orientation of the N- and C-terminal ends of the protein, as well as the central hydrophilic region, with respect to the cytoplasm (in) and cell exterior (out) (Figure 1A). Therefore, we aimed to reduce the number of likely topological scenarios by providing experimentally determined restrictions. Notably, all of the software predictions indicated one consistent feature, a long hydrophilic stretch in the PIN1 amino acid sequence positioned centrally between regions bearing the hydrophobic, presumptive TM domains (Figure 1B). We reasoned that determining the orientation of this hydrophilic loop would be a good starting point to discriminate between predictions and pave the way in resolving PIN1 topology.
Figure 1.
Transmembrane Topology Predictions of the PIN1 Protein.
(A) Distinct topologies for PIN1 as predicted by different software with detailed indication of respective TM domain organizations, each of them depicted by a rectangle that is color coded for hydrophobicity. All predictions indicate consistently a large hydrophilic region (depicted as a curved line; loop) of the protein that separates two TM domain regions (TMDs).
(B) The simplified 2D model of the protein shows the basic elements used in the initial topology determination stages, namely the TMDs separated by a central large hydrophilic loop.
The Hydrophilic Loop of PIN1 Localizes to the Cytoplasm
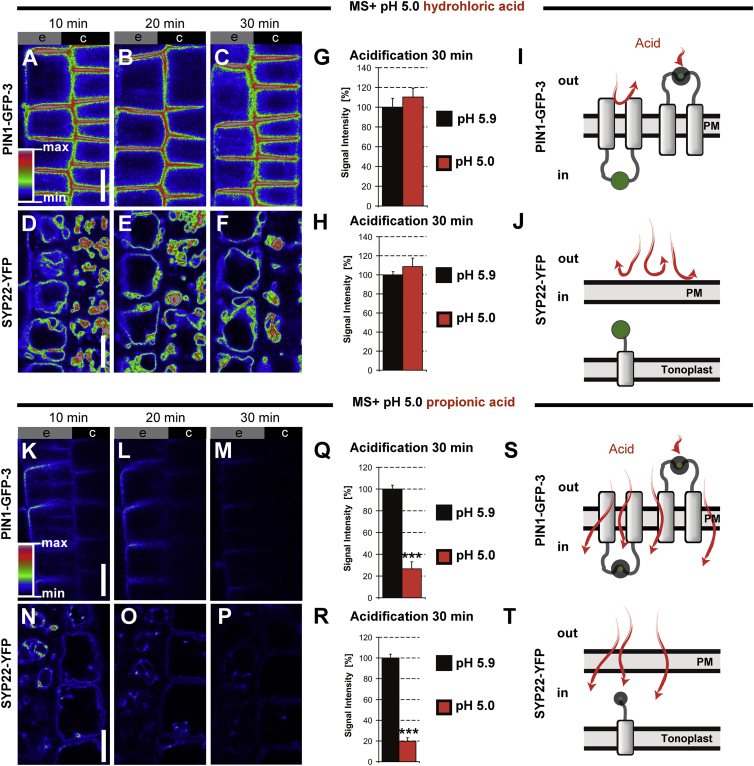
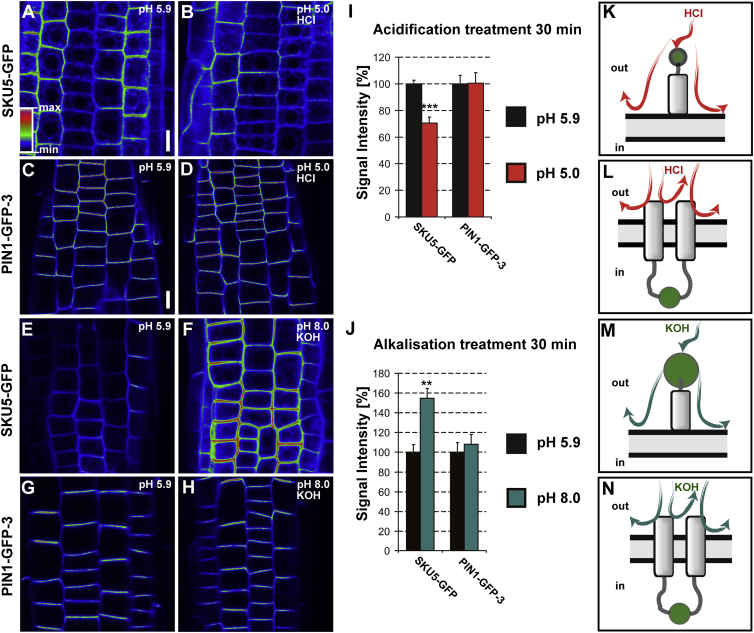
Previous biochemical and cell-biological studies have demonstrated that much of the information necessary for membrane trafficking and polar targeting is encoded in the amino acid sequence of the predicted HL of PIN proteins (Michniewicz et al., 2007, Huang et al., 2010, Zhang et al., 2010, Ganguly et al., 2014). Given that these signals depend on interaction with cytosolic kinases and phosphatases (Michniewicz et al., 2007), it is to be expected that the respective amino acids should be exposed to the cytosol. To experimentally ascertain whether the long central region of PIN1 is indeed facing the cytoplasm or, alternatively, cellular exterior, we used the PIN1-GFP-3 line bearing the GFP insertion between amino acids 421 and 422, thus placing it well within the hydrophilic loop (Wiśniewska et al., 2006). We exploited the pH sensitivity of fluorescent proteins (FPs) such as GFP or YFP (Kneen et al., 1998, Llopis et al., 1998, Domingo et al., 2010, Pogorelko et al., 2011) (see Supplemental Table 1). Thus, the fluorescence changes caused by acid treatment can provide information about the position of the FP relative to PM. We used the ionic acid HCl (membrane non-permeable) and the amphipathic propionic acid (membrane-permeable) to selectively acidify the apoplast or both the apoplast and cytoplasm, respectively. As a positive control for the intracellular acidification we used the prevacuolar compartment and vacuole marker SYP22-YFP, in which the YFP moiety is in the cell's interior and faces the cytoplasm (Robert et al., 2008). After treating PIN1:GFP-3 or SYP22-YFP for 30 min with HCl-titrated medium at pH 5.0, no fluorescence decrease of the respective FPs was observed (Figure 2A–2J). This suggests that both FPs were protected against the acid treatment by the presence of the PM. In contrast, when these lines were treated with medium buffered to pH 5.0 with membrane-permeable propionic acid (Figure 2K–2T), both PIN1-GFP-3 (Figure 2K–2M and 2Q) and SYP22-YFP (Figure 2N–2P) showed a gradual decrease of fluorescence, being very obvious over a 30-min treatment (Figure 2Q and 2R). This suggests that both FPs reside in the cytosol (Figure 2S and 2T). In addition, we tested another transgenic line designated PIN1-GFP-2 harboring the reporter inserted in the hydrophilic loop at the 217 amino acid position (see Figure 1A and Supplemental Figure 1A). This construct was also not affected by medium buffered with membrane non-permeable acid but was quenched by propionic acid-titrated medium (Supplemental Figure 1B). As an additional control, we treated analogically two transgenic lines expressing the YFP-AUX1 (N-terminally tagged with YFP) and AUX1-YFP-116 with the reporter moieties positioned in the cytoplasm, based on published results (Swarup et al., 2004). For both lines, the treatment with HCl-titrated medium (pH 5.0) did not quench the YFP while the propionic acid-titrated acted similarly as in the case of the PIN-GFP constructs (Supplemental Figure 1C and 1D). Next, we verified that the differences between fluorescence-quenching properties of the propionic acid versus HCl result from their different membrane permeabilities while their effect on GFP reporter quenching is specifically a result of lowering pH. We utilized a transgenic line expressing SKU5-GFP. As a glycosylphosphatidylinositol-anchored protein it is attached to the extracellular PM leaflet, so the fluorescent reporter is also facing the cellular exterior (Sedbrook et al., 2002, Mayor and Riezman, 2004). Treating SKU5-GFP with HCl-acidified medium (pH 5.0) for 30 min resulted in a decrease of its fluorescence in comparison with the control incubated in medium titrated to a standard pH of 5.9 (Figure 3A and 3B). In this setup, the PIN1-GFP-3 line, for which we determined the reporter location as cytoplasmic, was not affected by the treatment (Figure 3C and 3D). In addition, we performed analogical treatments on both marker lines, subjecting them to alkalization with medium titrated to pH 8.0 with membrane non-permeable base KOH. The results revealed an increased fluorescence of SKU5-GFP in comparison with control medium (Figure 3E and 3F) while the PIN1-GFP-3 was not affected (Figure 3G and 3H). The results were further supported by the quantitative analysis of acquired images, revealing statistically significant differences for both acidification and alkalization treatments (Figure 3I and 3J, respectively). These observations support the adequacy of our strategy and indicate that ionic, strong, fully dissociating acids or bases do not permeate the PM but affect the fluorescence of FPs in the apoplastic space (Figure 3K–3N). To confirm our results, we performed western blots with membrane protein fractions isolated from PIN1-GFP-3 and SKU5-GFP seedlings subjected to acid treatments as above (Figures 2 and 3) using an antibody against the GFP moiety. Neither for PIN1-GFP-3 nor SKU5-GFP did obvious degradation or protein level changes seem to be induced by HCl or propionic acid treatments, supporting the interpretation that the signal decrement was a result of pH-dependent quenching rather than protein degradation (Supplemental Figure 2).
Figure 2.
The Fluorescence of GFP Inside the Hydrophilic Loop of PIN1-GFP-3 Is Not Affected by Lowering the pH of the Extracellular Environment.
(A–F) Exchange of the growth medium (pH 5.9) by medium acidified to pH 5.0 with the membrane non-permeable hydrochloric acid (HCl) does not change the fluorescence intensity of PIN1-GFP-3 (A–C), or that of the vacuolar marker SYP22-YFP (D–F) in the root epidermis (e) and cortex (c). Scale bars, 10 μm.
(K–P) Exchange of the growth medium (pH 5.9) by medium acidified to pH 5.0 with the membrane-permeable propionic acid reduces the fluorescence of both PIN1-GFP-3 (K–M) and the vacuolar SYP22-YFP (N–P). Fluorescence intensities are color coded. Scale bars, 10 μm.
(G, H, Q, and R) Quantification of the fluorescence intensity changes recorded for the respective reporter lines after 30 min of acid medium exchange; 30 min of mock treatment is plotted as 100%. Error bars represent SE for three biological repeats (approximately 40 cells measured/seedling, number of seedlings imaged, n > 7 seedlings per biological repeat); ***P < 0.001.
(I, J, S, and T) Schemes illustrating the presumptive topological arrangements of PIN1-GFP-3 (I and S) and the known orientation of SYP22-YFP (J and T) with the reporter moiety indicated in green. For simplicity the TM regions of PIN1 are indicated only as two domains separated by the hydrophilic loop.
See also Supplemental Table 1 and Supplemental Figure 1.
Figure 3.
Acidification and Alkalization Treatment with Membrane Non-permeable Agents Respectively Decreases and Increases Fluorescence of Apoplastic GFP Moiety of SKU5-GFP.
(A–D) Exchange of the growth medium (pH 5.9) by medium acidified to pH 5.0 with membrane non-permeable hydrochloric acid (HCl) reduces the fluorescence of the extracellularly positioned SKU5-GFP (B) while having no pronounced effect on the signal of PIN1-GFP-3 (D) in comparison with the respective controls (A and C) in the root epidermis. Scale bars, 10 μm.
(E–H) Exchange of the growth medium (pH 5.9) by medium alkalized to pH 8.0 with membrane non-permeable potassium hydroxide (KOH) elevates the fluorescence of the extracellularly positioned SKU5-GFP (F) while having no significant effect on the signal of PIN1-GFP-3 (H) in comparison with the respective controls (E and G). Fluorescence intensities are color coded. Scale calibration as for (A)–(D).
(I and J) Quantification of fluorescence signal changes recorded for SKU5-GFP and PIN1-GFP-3 after treatment with medium buffered by HCl (I) or KOH (J) to pH 5.0 or pH 8.0, respectively; control treatment is plotted as 100%. Error bars represent SE for three biological repeats (number of seedlings imaged, n > 7 per each replicate); **P < 0.01, ***P < 0.001.
(K–N) Cartoons indicating the orientation of fluorescent reporters in the case of SKU5-GFP (K and M) and PIN1-GFP-3 (L and N) where the GFP moieties are facing the extracellular space or cytoplasm, respectively.
See also Supplemental Figure 2 and Supplemental Table 1.
Collectively those results show that the FP of PIN1-GFP-2 and -GFP-3, and thus the adjacent hydrophilic loop sequence, is positioned in the cytoplasm. These results thus allow us to discard topology software predictions 1, 2, and 6 (Figure 1A), which position the PIN1-HL (or the part of the loop fused to GFPs—amino acid positions 217 and 421 for GFP-2 and GFP-3, respectively) facing the apoplastic space.
The C-Terminal End of PIN1 Protein Faces the Apoplast
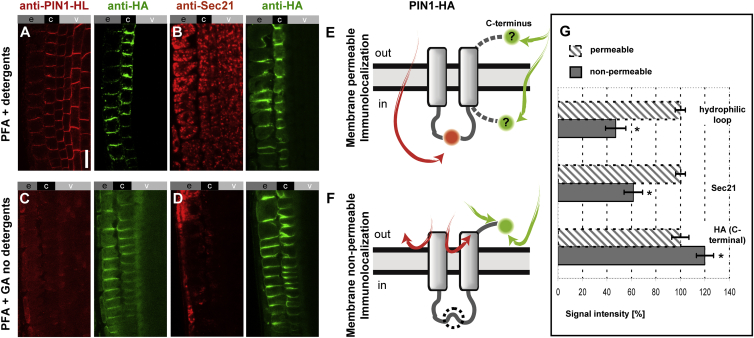
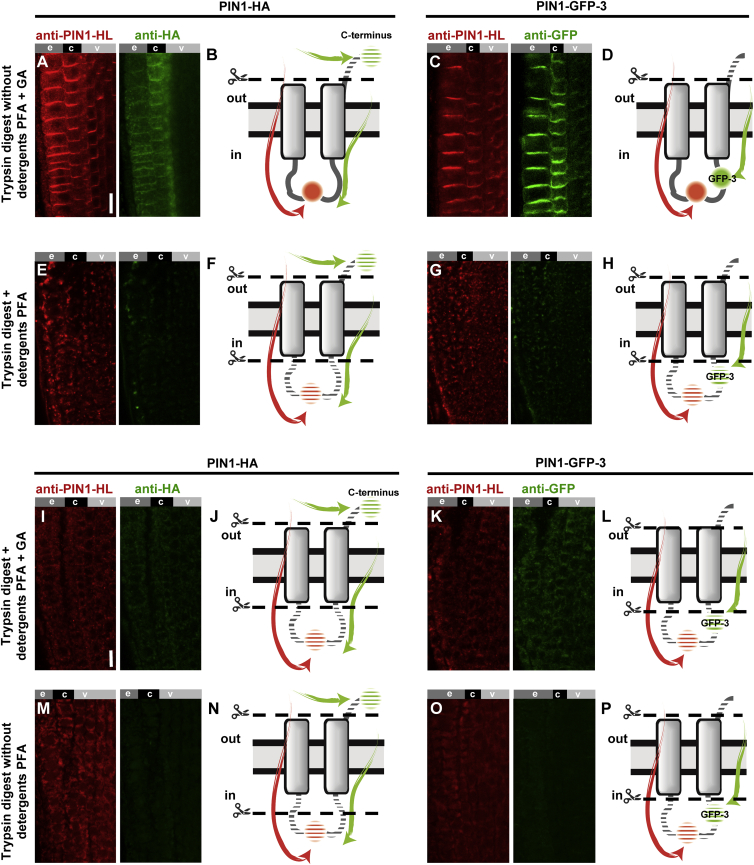
Among the remaining topological scenarios, a variation in the number of TM domains and orientation of the C-terminus of PIN1 is present (see Figure 1A, predictions 3, 4, and 5). Therefore, we next focused on resolving the orientation of the C-terminus of PIN1 using the functional C-terminally hemagglutinin (HA)-tagged PIN1 (PIN1-HA) (Wiśniewska et al., 2006). This construct allows simultaneous detection of the PIN1 hydrophilic loop using the anti-PIN1 polyclonal antibody (Paciorek et al., 2005) as well as the HA epitope using the anti-HA antibody. To be able to differentiate between extracellular and intracellular epitopes, we modified the standard immunolocalization protocol (Sauer et al., 2006) by excluding the lipid-disrupting agents at all steps. In addition, we also included glutaraldehyde (GA) in the fixation mixture to better preserve the integrity of lipids and, thus, the PM (Hopwood, 1972, Russell and Hopwood, 1976, Tanaka et al., 2010). When using the standard protocol, with paraformaldehyde (PFA) tissue fixation and utilization of detergents (Friml et al., 2003a, Sauer et al., 2006), the anti-PIN1 labeled both the PIN1-HA expressed ectopically in the epidermis and cortex and the endogenous PIN1 in the central tissues of the root (red, Figure 4A, left panel). The anti-HA labeled only PIN1-HA in the epidermis and cortex (green, Figure 4A, right panel). In a similar experiment using additional Golgi-specific anti-Sec21 antibody (Movafeghi et al., 1999, Pimpl et al., 2000) we observed, besides the HA labeling (Figure 4B right panel, green), also clear intracellular Golgi labeling (visible in red, Figure 4B, left panel), confirming that in this protocol the antibodies permeate the PM (Figure 4E). Next, we used the same antibodies in an immunolocalization protocol whereby PFA and GA fixation was utilized and detergents were excluded to reduce membrane permeation to antibodies. For brevity, we refer to this protocol as “membrane non-permeable,” whereas the standard protocols referred to as “membrane-permeable”. When using the membrane non-permeable conditions for immunolocalization, we observed a strongly decreased labeling with anti-PIN1 (Figure 4C, left panel, red) while the HA signals remained clearly visible (Figure 4C, right panel, green). Similarly, the Golgi marker anti-Sec21 (visible in red, Figure 4D, left panel) exhibited a markedly weaker labeling while the HA signal persisted (Figure 4D, right panel, green). These observations show that under these conditions membrane integrity was sufficiently preserved, limiting the permeabilization of antibodies to the cell interior (Figure 4F). The results were supported by quantifications of signals corresponding to PIN1-HL and Sec21 that were significantly more intense in the membrane-permeable protocol (Figure 4G, striped bar) compared with the non-permeable conditions (solid bars). Notably, PIN1-HA labeling was even stronger in non-permeable conditions, possibly resulting from higher anti-HA antibody availability when it did not penetrate to the cell interior, thus being more available to target the extracellular epitopes (Figure 4G).
Figure 4.
The C-Terminally Positioned HA Tag in PIN1-HA Is Stained Preferentially, Over the Hydrophilic Loop, in Membrane Non-permeable Conditions, Indicating Its Extracellular Location.
(A–D) Double antibody staining of PIN1-HA using anti-HA in combination with anti-PIN1-HL (A and C), or with anti-Sec21 (B and D) in membrane-permeable (A and B) and non-permeable (C and D) conditions; the labeled epitopes are expressed in epidermis (e), cortex (c), and vasculature (v) of Arabidopsis seedling roots. Scale bar, 10 μm.
(E and F) Interpretative cartoons indicating the orientation of hydrophilic loop and the C-terminal HA tag in PIN1 and the results of their dual immunostaining in membrane-permeable (E) versus non-permeable (F) conditions. The designations “in” and “out” refer to the intracellular and the extracellular space, respectively. The dashed circle (F) represents absence of PIN1-HL labeling.
(G) Signal quantification of the HA, Sec21, and PIN1 loop labeling plotted as a comparison of membrane-permeable versus non-permeable immunolocalization conditions; immunolocalization in membrane-permeable conditions is plotted as 100%. Error bars represent SE for three biological repeats (number of seedlings imaged, n > 5 per each replicate). *P < 0.05.
See also Supplemental Figure 3.
We performed additional experiments to verify whether removal of detergents alone and PFA fixation is sufficient for membrane non-permeability. Such a protocol resulted in pronounced PIN1-HL, Sec21, and HA signals (Supplemental Figure 3A–3D), indicating that membrane integrity was not sufficiently preserved. However, in such conditions labeling of PIN1 and the Golgi marker was not as uniform throughout the root as in standard conditions, indicating that some permeability limitations occurred. Similarly, the protocol employing PFA and GA fixation but with addition of detergents resulted in decoration of all of the epitopes of interest, similar to a standard permeable immunolocalization protocol (see Figure 4A and 4B).
To corroborate these observations, we performed analogical immunolocalizations on AUX1-YFP-116 and AUX1-HA transgenic lines both harboring tags (YFP and HA) facing the intracellular space (Swarup et al., 2001, Swarup et al., 2004). Standard protocol (PFA, detergents) used on the AUX1-YFP-116 transgenic line resulted in labeling of YFP with anti-GFP antibody (red, Supplemental Figure 3I) and with the signal of YFP itself remaining fluorescent after fixation (green, Supplemental Figure 3J). Similarly anti-HA (green, Supplemental Figure 3K) and anti-PIN1-HL (red, Supplemental Figure 3L) signals were well visible in the N-terminally tagged HA-AUX1 line. In contrast, immunolocalizations in membrane non-permeable conditions (PFA, GA, no detergents) resulted in visibly weaker labeling of intracellularly localized epitopes targeted by anti-GFP, anti-HA, and anti-PIN1-HL antibodies (Supplemental Figure 3M–3P). Similar results were obtained with the PIN1-GFP-2 transgenic line, showing anti-GFP antibody labeling only when using the membrane-permeable protocol (Supplemental Figure 2R–2U) and thus confirming that the middle loop is located intracellularly. These results indicate that both modifications, i.e., removal of detergents and utilization of the GA as additional fixative, contribute to preferential labeling of extracellular epitopes, likely due to greater preservation of PM integrity. Collectively, these data confirm that the middle hydrophilic loop is intracellular and reveal that the C-terminus of PIN1 is facing the extracellular space.
A Protease Protection Assay Confirms the Apoplastic Orientation of the PIN1 C-Terminus
To further verify our findings and expand the palette of techniques that can be used to study the topology of proteins in plants, we decided to test and adapt a version of the protease protection assay (Lorenz et al., 2006) for plant material. We took advantage of PIN1-HA-expressing seedlings, subjecting them to immunolocalization in non-permeable conditions. This time, however, we introduced a trypsin digestion step to the protocol before applying antibodies to fixed seedlings. Subsequent imaging revealed that the PIN1-HL signal was preserved (Figure 5A, left panel, red) while the HA was distinctly weaker (Figure 5A, right panel, green). The presence of the red signal corresponding to the intracellular PIN1-HL was initially surprising, as no detergents were included in the procedure. Possibly the tryptic digests of protein complexes at the PM would result in the formation of pores that are large enough for the antibodies to pass through at the following stages of the protocol (Figure 5B). In addition, we used the PIN1-GFP-3 line treated in the same fashion, which resulted in preservation of the epitopes localized on the hydrophilic loop (anti-PIN1-HL) as well as the intra-loop GFP-3 moiety that was labeled with anti-GFP (Figure 5C and 5D). We also tested the intracellular markers SYP22-YFP (tonoplast) and Sec21 (Golgi), which consistently were protected from trypsin, but labeled respectively by anti-GFP and anti-Sec21 (Supplemental Figure 4A–4D), indicating that the trypsin treatment enables antibodies to permeate to the intracellular space at subsequent stages of the immunocytochemistry protocol. Utilization of an analogical setup but in membrane-permeable conditions (PFA fixation and detergents) resulted in disappearance of the characteristic polar signal of anti-PIN1-HL as well as of anti-HA, indicating digestion of both epitopes (Figure 5E and 5F). A similar result was observed for the PIN1-GFP-3 using anti-GFP (Figure 5G and 5H). Congruently, in those conditions the epitopes within intracellular markers SYP22-YFP and Sec21 targeted respectively by anti-GFP and anti-Sec21 antibodies were also destroyed by trypsin digestion, resulting in a lower and more diffuse signal (Supplemental Figure 4E–4H).
Figure 5.
Limited Membrane Permeability Prevents Tryptic Digestion of Intracellular Epitopes of PIN1-HA and PIN1-GFP-3.
(A, C, E, and G) Antibody staining of PIN1-HA (A and E) and PIN1-GFP-3 (C and G) after tryptic digest in membrane non-permeable (A and C) versus membrane-permeable (E and G) conditions. Scale bar, 10 μm.
(B, D, F, and H) Interpretative cartoons depicting the presumptive orientations of epitopes in PIN1 hydrophilic loop (PIN1-HL; red) and the C-terminal HA tag (green) in PIN1-HA (B and D) as well as the internal GFP in PIN1-GFP-3 (F and H). Arrows indicate the accessibility of respective epitopes to anti-PIN1-HL (red), anti-HA (green) and anti-GFP (green) antibodies. Shaded areas reflect destruction of the epitopes after tryptic digest in membrane non-permeable (B and D) versus permeable (F and H) conditions. The designations “in” and “out” refer to the intracellular and the extracellular space, respectively.
(I, K, M, and O) Antibody staining of Arabidopsis PIN1-HA (I and M) and PIN1-GFP-3 (K and O) fixed with paraformaldehyde (PFA) and glutaraldehyde (GA) mixture but permeabilized with detergents prior to tryptic digest (I and K) and analogically treated samples fixed only with PFA without detergents used (M and O). Scale bar, 10 μm.
(J, L, N, and P) Interpretative cartoons depicting the presumptive orientations of epitopes in PIN1 hydrophilic loop (PIN1-HL; red) and the C-terminal HA tag (green) in PIN1-HA transgenic line (J and N) as well as the internal GFP in PIN1-GFP-3 transgenic line (L and P). Arrows indicate the accessibility of respective epitopes to anti-PIN1-HL (red), anti-HA (green), and anti-GFP (green) antibodies. Shaded areas reflect destruction of the epitopes due to tryptic digest.
See also Supplemental Figure 4.
When analogical experiments were performed either with PFA and GA fixation but including detergents or PFA fixation alone, tryptic digests of all epitopes of interest were observed (Figure 5I–5P). These results supported the notion that use of GA as fixative with simultaneous omission of detergents contributes to preferential digestion of extracellular epitopes and protection of those positioned in the cytoplasm, making it inaccessible to trypsin. In addition, the thorough digestion of proteins across the root tissues (Figure 5I–5L) illustrates that the activity of trypsin was not diminished by residual fixative (GA or PFA) that has been thus successfully removed and inactivated during tissue post-fixation steps (see Methods). Also, under our conditions, the proteins are not excessively crosslinked to be rendered indigestible (Bliss and Novy, 1899, Webster et al., 2009). Altogether, these results give credence to the proposal that in our protocols the particular results observed are due to effects on membrane permeability rather than masking of epitopes. With those two approaches, i.e., membrane-permeable versus non-permeable immunolocalization and application of trypsin in this setup, we provide strong evidence for the extracellular position of the C-terminus of PIN1 and intracellular location of its hydrophilic loop (also revealed by the pH manipulation). This allows us to discard prediction number 3 that indicates the cytoplasmic position of the C-terminus (Figure 1A) and to further narrow the possible topologies of PIN1.
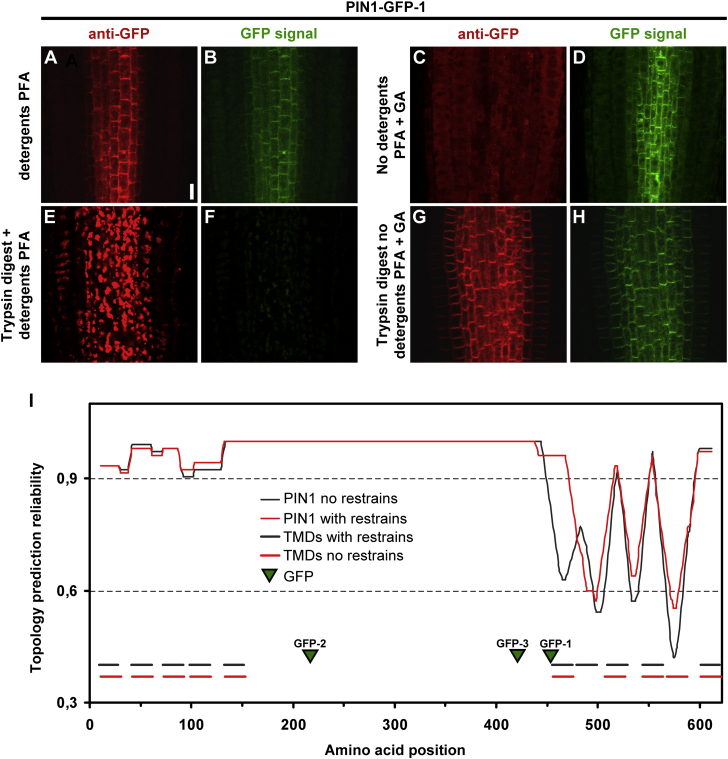
The GFP-1 Reporter Fusion in PIN1 Exhibits Features of Both Intra- and Extracellular Location
To further refine our understanding of PIN1 topology, we investigated PIN1-GFP-1 (Benková et al., 2003), which has the GFP inserted in the area where different predictions disagree about the possible end of the hydrophilic loop (GFP positioned between amino acids 453 and 454; see Figure 1A). We subjected this line to a standard immunolocalization, labeling the GFP with anti-GFP antibody (Figure 6A, red) while also directly visualizing the native GFP fluorescence (Figure 6B, green). An analogical but membrane non-permeable protocol resulted in the absence of the anti-GFP signal while retaining GFP fluorescence (Figure 6C and 6D). As described earlier, a membrane-permeable immunolocalization preceded by trypsin treatment resulted in disappearance of the characteristic polar, PM PIN1 localization with both red and green signals becoming more diffuse, consistent with a proteolytic cleavage (Figure 6E and 6F). When trypsin was used in a membrane non-permeable protocol, PIN1-GFP was not visibly affected (Figure 6G and 6H). In addition, we subjected the line to acidification and alkalization treatments as described above (see Figure 3). These revealed a noticeable decrease of fluorescence after 30 min of incubation with HCl-titrated medium at pH 5.0 (Supplemental Figure 5A and 5B), suggesting extracellular GFP localization. However, the alkalization treatment with KOH-titrated medium at pH 8.0 did not visibly affect the signal of PIN1-GFP-1 (Supplemental Figure 5C), hinting again toward intracellular localization. This is in direct contrast to the acidification results using PIN1-GFP-2 or PIN2-GFP-3. However, these PIN1-GFP constructs are expressed in a tissue context (epidermis, cortex) different from that of PIN1-GFP-1 (stele, endodermis). Therefore, we tested analogically the PIN1::PIN1-YFP line that has reporter fusion with a comparable pH sensitivity (see Supplemental Table 1), exactly at the same position as the intracellularly located PIN1-GFP-2 (amino acid 217). Consistently in our results using PIN2::PIN1-GFP-2 (Supplemental Figure 1B), no pronounced fluorescence intensity differences of the YFP after both acidification and alkalization treatment could be observed (Supplemental Figure 5D–5F). Quantification of PIN1-GFP-1 confocal images confirmed a statistically highly significant quenching of GFP-1 signal after acidification but not after alkalization (Supplemental Figure 5G), whereas the PIN1::PIN1-YFP line expectedly did not show any statistically significant signal changes after either treatment (Supplemental Figure 5H).
Figure 6.
Immunolocalization Protocols Indicate Intracellular Position of the Reporter Moiety in PIN1-GFP-1.
(A and B) Immunolocalization protocol, performed on PIN1-GFP-1-expressing roots, in membrane-permeable conditions results in labeling of the GFP with anti-GFP antibody (red; A) while the fluorescent signal of GFP alone is also visible (green; B).
(C and D) PIN1-GFP-1 immunolabeling in membrane non-permeable conditions results in absence of anti-GFP labeling (red signal not visible; C) while the native fluorescent signal of GFP remains visible (green; D).
(E and F) Antibody staining after tryptic digest in membrane-permeable conditions results in disappearance of the polar PM signal of immunolabeled anti-GFP (red; E) as well as green fluorescence of the reporter alone (green; F) due to proteolytic degradation.
(G and H) Analogical antibody staining after tryptic digest in membrane non-permeable conditions results in partial GFP-1 immunolabeling (red; G) with the green fluorescence of the reporter (green; H) intact. Scale bar in (A) represents 10 μm (applies to A–H).
(I) Plot of topology prediction reliability in two variants: the black line indicates the reliability of de novo topology prediction without constraints. The red plot indicates the topology prediction reliability when constraints derived from our empirical data are imposed: the GFP insertions 2 and 3 are positioned in the cytoplasm while the GFP-3 is indicated as positioned in the membrane; the C terminus is out. Depicted on the bottom is the topology prediction of PIN1 TM domains (depicted as lines) and the position of GFP insertions (green arrowheads) in the PIN1-GFP.
See also Supplemental Figures 5 and 6.
In summary, all our methods, while working for other epitopes and GFP positions, show somewhat ambivalent results specifically for the GFP-1 position, exhibiting some features of both intra- and extracellular locations. This suggests that the GFP might be in a position where it interferes with PIN1 functioning. However, PIN1-GFP-1 fully complemented the characteristic inflorescence phenotype in the sterile pin1-201 null allele, and did not induce other floral aberrations when expressed in wild type Col-0 (Supplemental Figure 6A and 6B). This suggests that the GFP-1 insertion does not greatly impair PIN1-GFP-1 function and that this transgenic line can be used in topology studies.
Topological Modeling Supported by Empirical Data Hints at 10-TM Domain Topology of PIN1
Armed now with a wealth of empirical data on topological positions of different parts of the PIN1 protein, we utilized TOPCONS, a software suite that generates a consensus topology for a target protein and allows input of empirical data to enhance the predictions, while also plotting the calculated prediction reliability for particular protein regions (Bernsel et al., 2009). TOPCONS processing of a PIN1 sequence without any empirical constraints produced a 10-TM domain protein model (see horizontal lines in Figure 6I at bottom of the graph) based on the agreement level of several underlying topology predictors. The prediction reliability was reasonably high for the first bundle of helices (TM domains 1–5); however, the prediction was unreliable for the second group (TM domains 6–10; see Figure 6I, black line plot). Subsequently, we added the constraints derived from our empirical data. We designated the topological arrangement of PIN1 amino acids in insertion regions of GFP-2 and GFP-3 as intracellular and the position of PIN1 C-terminus as extracellular. However, the position of the GFP-1 was conceptually more challenging to classify. The immunolocalization and trypsin digestion data indicated an intracellular position of the GFP-1 (Figure 6A–6H) while the acidification experiments showed quenching of that reporter by non-permeable acid, indicating its extracellular localization (Supplemental Figure 5A–5C and 5G). The unrestrained TOPCONS model indicated the GFP-1 as inside but in fact touching the membrane just before the beginning of the sixth α helix with one amino acid slack from the bilayer. Regarding also the experimental ambiguities, we assumed the GFP-1 position as dipped in the membrane from the inside. The resulting 10-TM model showed a location change for two predicted helices (see horizontal red lines in Figure 6I at the bottom of the graph). The prediction reliability for the bundle of helices proximal to the C-terminus of the protein (TM domains 6–10) was increased overall (see Figure 6I, red plot). Notably, we observed higher reliability only with GFP-1 position indicated as membrane, while designating it as fully out or in yielded lower reliability (data not shown). Gathering all the evidence, we can conclude that PIN1 is a polytopic protein that is most likely composed of 10 TM domains with an intracellular central loop and extracellular termini.
Plasma Membrane-Localized PINs Likely Share a Similar Topology
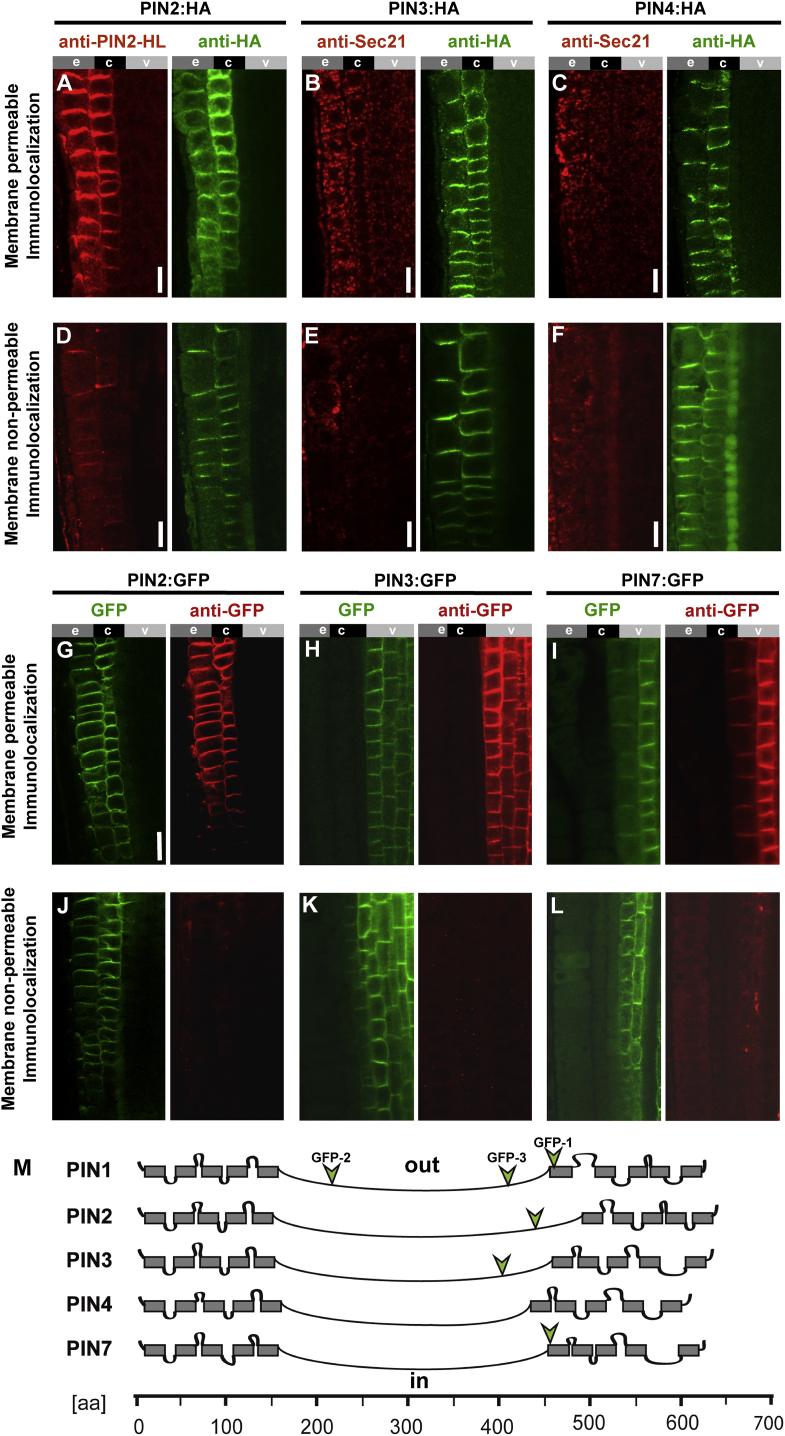
Taking advantage of the available Arabidopsis transgenic lines expressing tagged versions of PM-localized PINs, we subjected the C-terminally HA-tagged PINs 2, 3, and 4 (Wiśniewska et al., 2006) to the membrane-permeable and non-permeable immunolocalization protocols described above. The HA was labeled both in membrane-permeable and non-permeable conditions while the HL of PIN2, targeted by the anti-PIN2(HL) antibody, as well as the intracellular marker Sec21, were visibly less labeled when membrane permeability was limited (Figure 7A–7F). This suggests that PIN2, PIN3, and PIN4 also have intracellularly positioned hydrophilic loops and extracellular C-termini. We also tested intragenically GFP-tagged PIN2, PIN3, and PIN7 proteins. The fluorescent reporters in PIN2-GFP (Xu and Scheres, 2005) and PIN3-GFP (Žádníková et al., 2010) are inserted inside the hydrophilic loop in locations topologically proximal to tested PIN1-GFP-3 that was determined as intracellular. The GFP in PIN7-GFP (Blilou et al., 2005) is located similarly as in PIN1-GFP-1 at the very end of the hydrophilic loop (for GFP positions, see Figure 7M). In all of these cases, the anti-GFP immunolocalization revealed a visibly less pronounced immunolabeling in membrane non-permeable conditions (Figure 7G–7L). The results are entirely congruent with those obtained for PIN1. In summary, the results suggest a similar 10-TM domain topology for all Arabidopsis PM-localized PINs. This is likely a result of a high degree of sequence conservation in the helical regions among the subclade of the so-called long PIN proteins (Supplemental Figure 7).
Figure 7.
The C Terminus Is Oriented Extracellularly in Other PM PINs.
(A–C) Antibody staining of PIN2-4-HA in membrane-permeable conditions results in a pronounced labeling (red) of the hydrophilic loop of PIN2 (PIN2-HL; A) and the cytoplasm-located Sec21 (B and C) as well as the C-terminally positioned HA tag (green); labeled epitopes are expressed in the epidermis (e), cortex (c), and vasculature (v) of root. Scale bars, 10 μm.
(D–F) Immunolocalization in membrane non-permeable conditions results in pronounced HA signal (green right-hand panels) and a markedly weaker labeling of the intracellular marker Sec21 (red; E and F) as well as a weaker labeling of the hydrophilic loop (red; D) indicating its intracellular localization. Scale bars, 10 μm.
(G–I) Antibody staining of PIN2, PIN3 and PIN7 transgenic lines in membrane-permeable conditions results in a pronounced anti-GFP labeling (red) while the fluorescence of GFP itself remains visible (green) in the epidermis (e), cortex (c), and vasculature (v) of root. Scale bar in (G) represents 10 μm (applies to G–L)
(J–L) Immunolocalization in membrane non-permeable conditions results in absence of the red signal corresponding to GFP antibody labeling (right-hand panels) while the green fluorescent signal of GFP itself remains visible.
(M) Schematic topology representation of PM-localized PIN1–4 and PIN7 modeled with empirical constraints. The TM domains are depicted as rectangles. All predictions indicate consistently a large hydrophilic region between the TM domain bundles. The positions of GFP insertions are indicated with green arrowheads. Note that the position of YFP in PIN1::PIN1-YFP is the same as the GFP-2 position indicated above.
See also Supplemental Figure 7.
Discussion
Methods for Topology Determination of Plant PM Proteins: Expanding the Toolkit
The toolkit for topology studies in plant cells is still relatively limited and mostly based on FPs (Ohad et al., 2007, Brach et al., 2009, Osterrieder et al., 2009, Sparkes et al., 2010). For topology mapping of PM-localized PINs, we elaborated on the FP pH sensitivity principle previously used, for example, in AUX1 IAA−/H+ symporter topology study (Swarup et al., 2004). The fluorescence of FPs decreases in line with lowering pH, thus allowing deduction of their position relative to the PM. This method is straightforward, as it can be performed on available reporter lines originally generated for other purposes such as cell biology investigations. We have demonstrated that the usually low fluorescence of the extracellularly positioned reporter, SKU5-GFP (Gjetting et al., 2012), can be made brighter by elevating the pH of the extracellular environment. In contrast, changing the extracellular pH did not affect the fluorescence intensity of intracellular FPs, e.g., the tonoplast marker SYP22-YFP. Thus, we validated the use of this acid sensitivity assay for topology mapping of extracellularly versus intracellularly positioned FPs.
Unfortunately, GFP tagging of the PIN1 N- or C-terminus for topology studies was not possible because none of the terminal fusions with a bulky GFP ever yielded a significant fluorescence, a difficulty also encountered during AUX1 C-terminal tagging attempts for topology determination (Swarup et al., 2004). However, it was possible to C-terminally tag PIN1 with a smaller epitope, such as HA, that did not interfere with PIN1 functionality (Wiśniewska et al., 2006). Therefore, we developed a modified immunolabeling procedure to selectively detect extracellular epitopes. The key to this procedure was to limit PM permeability during fixation by supplementing the standard fixative with GA. This potent protein crosslinker, more reactive than PFA (Sabatini et al., 1963, Migneault et al., 2004), is also able to react with some phospholipids (Hopwood, 1972, Russell and Hopwood, 1976, Tanaka et al., 2010). Therefore, the limited PM permeability impedes the penetration of antibodies to the cell's interior, resulting in preferential labeling of extracellularly exposed epitopes. Using the modified tissue fixation protocol, we further corroborated the extracellular position of the C-terminal HA tag in PIN1-HA via a protease protection assay (Lorenz et al., 2006) that we adapted for use in a plant whole-mount immunolocalization protocol.
Thus, we successfully adapted several methods that allowed us to probe in planta the extra- and intracellular positions of different parts of PM proteins. Such experimental output, in combination with publicly available online topology predictions software, constitutes a powerful pipeline for rapid topology determination of PM proteins in plants.
Topology Modeling: Shaping the Questions and Guiding the Experimental Efforts
In this study, we investigated multiple bioinformatics predictions to obtain an overall view of possible PIN1 topologies. Consensus (Nilsson et al., 2000) and so-called partial consensus topologies approaches (Nilsson et al., 2002) have been used to reconcile multiple possible topologies, a strategy intended to minimize experimental efforts, focusing them specifically on the protein parts with the poorest consistency among the predicted topologies. Further software developments in the field implemented the use of limited experimental data to markedly improve the reliability of topology models (Drew et al., 2002, Kim et al., 2003, Rapp et al., 2004, Daley et al., 2005). In the case of PIN proteins, models agree strongly about the N-terminal bundle of TM domains, also somewhat complementing the need for experimental efforts that would have to be otherwise devoted to this region which, due to the past multiple unsuccessful N-terminal tagging attempts of PIN1, would be technically demanding. Thus, we focused our experimental efforts on defining the position of the central hydrophilic loop and the C-terminal group of helices. To take the optimal advantage from the approaches delineated above we used the TOPCONS software suite (Bernsel et al., 2009), which is capable of generating consensus predictions that can be enhanced by experimental constraints. Feeding PIN1 topology empirical data into TOPCONS produced an increased prediction certainty for the second α-helical bundle. The prediction for N-terminal helices and the cytoplasmic hydrophilic loop remained highly certain, plotting a final 10-TM domain topology for PIN1 with the proteins' termini facing the apoplastic space. TOPCONS also indicated that the GFP in PIN1-GFP-1 is dipped in the membrane from the cytoplasmic side, suggesting that the unexpected HCl sensitivity of this reporter is a result of this somewhat unusual topological arrangement. It is also interesting to speculate whether the topology of PIN1 is entirely stable in the region of GFP-1 insertion and whether the reporter does not slip in and out of the membrane, therefore exhibiting its acid sensitivity. Indeed, the topology prediction programs indicate more variability in that particular region of PIN1 (Figure 1A). There are reports of proteins with an undefined, alternating topology (Bowie, 2006, Rapp et al., 2006). Similarly, the arrangement of TM domains in the lipid bilayer can be more complicated, including longer TM domains crossing the PM at an angle. In addition, so-called disrupted helices have been reported. These exhibit a break in α-helical structure continuity in the middle of the membrane (Elofsson and von Heijne, 2007). On the other hand, the GFP-1 insertion does not abolish PIN1 functionality, as we have shown (Supplemental Figure 6), indicating that the protein structure likely is not severely altered with the overall native topology preserved.
However, fully resolving those questions encourages and requires more detailed structural studies in the future.
Evolutionarily Conserved Topology of PM-Localized PIN Proteins
Arabidopsis PINs can be divided in the PM-localized long (harboring the long HL) PINs (PIN1–4 and 7) and the short ER-localized ones (PIN5, 8) as well as PIN6, which has an HL of intermediate length (Viaene et al., 2013). In general the TM domains of the long PINs are much more conserved than the HL (Křeček et al., 2009). Interestingly, a large-scale sequence alignment of multiple PIN HL sequences from land plants reveals some HL conserved regions, many of which contain known phosphorylation sites (Bennett et al., 2014) regulating PIN function (Ganguly et al., 2012). These motifs have to be accessible to cytoplasmic enzymes that modify the phosphorylation status (Michniewicz et al., 2007, Zhang et al., 2010, Barbosa et al., 2014, Zourelidou et al., 2014) and conformation (Xi et al., 2016) of the HL, supporting its intracellular localization also in congruence with our findings for Arabidopsis PM PINs. This also illustrates the adequacy of choosing the HL as the first protein region to be topologically mapped, as its likely intracellular orientation was a good validation of our experimental methods.
As mentioned above, the TM domains of PM-localized Arabidopsis PINs exhibit a high degree of sequence conservation, so a similar 10-TM domain topology among PINs is likely and supported by our empirical data for PINs 1–4 and PIN7. Moreover, the TM domains of canonical PINs seem to be conserved among land plants in general (Viaene et al., 2013, Bennett et al., 2014), hinting at their common topology. In addition, published alignments of multiple long PIN sequences indicate that the minor loops connecting the helices also harbor some highly conserved residues, hinting at their regulatory role (Bennett et al., 2014). This might explain the differences in the ability to become organized in polar domains between the endogenous PIN2 and the chimeric version of PIN5 (natively a short type PIN) containing the loop of PIN2 (PIN5:PIN2-HL) that can reach the PM (Ganguly et al., 2014). While the transfer of HL was sufficient to mediate trafficking to the PM, it could not achieve the characteristic polar localization and apoplast attachment seen for PIN2. This suggests that polarity and cell wall attachment are not only defined by features encoded in the HL of the long PINs, but also involve TM domains and their minor loops (see also Feraru et al., 2011). Therefore, the empirically verified and enhanced topological model should be instructive in elucidating the structure–function relationships of distinct PIN protein regions, constituting a road map for further structural investigations.
Methods
Growth Conditions
Seedlings for acid treatments and immunocytochemistry were grown vertically in Petri dishes on 0.8% agar 0.5× MS medium containing 1% sucrose (pH 5.7) at 18°C and under long-day photoperiod.
Plant Material
PIN2::PIN1-GFP2, PIN2::PIN1-GFP3, PIN2::PIN1-HA, PIN2::PIN3-HA, PIN2::PIN4-HA (Wiśniewska et al., 2006); AUX1::AUX1-YFP N-terminal, AUX1::AUX1-YFP 116 (Swarup et al., 2004); AUX1-HA (Swarup et al., 2001, p.2001); SKU5::SKU5-GFP (Sedbrook et al., 2002); PIN2::PIN2-HA (Vieten et al., 2005); PIN3::PIN3-GFP (Žádníková et al., 2010); PIN1::PIN1-GFP-1 (Benková et al., 2003); PIN1::PIN1-YFP, PIN2::PIN2:GFP (Xu et al., 2006); PIN7::PIN7-GFP (Blilou et al., 2005); SYP22::SYP22-YFP (Robert et al., 2008); pin1-201/+ SALK_047613 line (Alonso et al., 2003, Furutani et al., 2004) was genotyped using primer information deposited in the SALK database (http://signal.salk.edu/tdnaprimers.2.html); pin1-201 × PIN1-GFP-1 was made by genetic crosses and genotyped to obtain a homozygous line for both insertions.
Acid Treatments
For acid and base incubations, 5-day-old-seedlings were used; for membrane non-permeable acid-quenching experiments, MS liquid medium (pH 5.0) was titrated with HCl; for the membrane-permeable acid-quenching experiments, MS medium buffered to pH 5.0 containing 20 mM propionic acid was used; for membrane non-permeable base treatments, MS liquid medium (pH 8.0) titrated with KOH was used.
Tissue Fixation
For membrane-permeable immunocytochemistry protocols, Arabidopsis seedlings were fixed in 4% paraformaldehyde dissolved in PBS buffer. For membrane non-permeable immunocytochemistry protocols, a mixture of 4% paraformaldehyde and 0.025% GA dissolved in PBS buffer (pH 7.4) was used. Glutaraldehyde 25% stock was always added freshly to a pre-prepared 4% paraformaldehyde solution. In both protocols tissues were fixed for 45 min under vacuum. After fixation the residual non-reacted GA was inactivated by incubation of seedlings for 1 h in a freshly made 0.1% solution of sodium borohydride (NaBH4) dissolved in PBS (pH 7.4). Seedlings were then washed three times with 1 ml of PBS and subjected to the immunocytochemistry protocol.
Immunocytochemistry
Whole-mount in situ immunocytochemical protein localization in membrane-permeable conditions was performed as previously published (Sauer et al., 2006). For the immunocytochemical protein localization in membrane non-permeable conditions the same procedure as published previously was used (Sauer et al., 2006) except that all detergents and DMSO were removed from the protocol. For HA-tag detection, anti-HA raised in mouse antibody (Abcam) was used (1:600) labeled with a secondary antibody (Alexa Fluor 488, dilution 1:600). For labeling of PIN1 hydrophilic loop, a specific rabbit antibody raised against this protein part was used (Friml et al., 2003b) in 1:900 dilution and subsequently labeled with CY3 secondary antibody (1:600; Sigma-Aldrich). For labeling of GFP, an anti-GFP (1:600; Sigma) antibody raised in mouse was used. For labeling of the Sec21 Golgi-localized marker, a specific anti-Sec21 antibody (Movafeghi et al., 1999) raised in rabbit diluted 1:800 was used and subsequently labeled with CY3 secondary antibody (1:600; Sigma-Aldrich). Immunocytochemistry was performed using the InsituPro VSi system manufactured by INTAVIS Bioanalytical Instruments.
Confocal Image Acquisition and Fluorescent Signal Analysis
Due to signal intensity differences between fluorescent reporters, the confocal microscope settings was varied accordingly to satisfy optimal dynamic range for intensity measurement and facilitate the acquisition of quality pictures. To enable accurate quantification of signal changes, we match a particular sample with its untreated control imaged in one session during which the same confocal settings are held for the sample and control. The signals are then represented as percentages, allowing for comparison of different fluorescent reporters and biological repeats. Signal intensities were measured using Fiji software (https://fiji.sc).
Images were collected using Leica SP2 or Carl Zeiss 780 and 700 confocal microscopes. Figures were processed and assembled in Photoshop and InDesign CS5 (Adobe Systems).
Western Blot
For western blot experiments after acid treatment, the membrane protein fraction containing PIN1-GFP-3 protein loaded on gel was obtained from 25 mg of starting material (5-day-old seedlings); for SKU5-GFP, 13 mg was used. Membrane protein fractions were isolated using the previously published protocol (Abas et al., 2006).
Funding
This research has been financially supported by the Ministry of Education, Youth and Sports of the Czech Republic under the project CEITEC 2020 (LQ1601) (T.N., M.Z., M.P., J.H.), Czech Science Foundation (13-40637S [J.F., M.Z.], 13-39982S [J.H.]); Research Foundation Flanders (Grant number FWO09/PDO/196) (S.V.) and the European Research Council (project ERC-2011-StG-20101109-PSDP) (J.F.).
Author Contributions
T.N., M.Z., and M.P. performed the research and analyzed the data. M.P. and J.H. additionally contributed to data analysis and statistical quantifications. T.N., S.V., and J.F. designed the research and wrote the paper with input from all authors.
Acknowledgments
We thank David G. Robinson and Ranjan Swarup for sharing published material; Maria Šimášková, Mamoona Khan, Eva Benková for technical assistance; and R. Tejos, J. Kleine-Vehn, and E. Feraru for helpful discussions. The authors declare no conflict of interest.
Published: September 9, 2016
Footnotes
Published by the Molecular Plant Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, SIBS, CAS.
Supplemental Information is available at Molecular Plant Online.
Contributor Information
Tomasz Nodzyński, Email: tomasz.nodzynski@ceitec.muni.cz.
Jiří Friml, Email: jiri.friml@ist.ac.at.
Supplemental Information
References
- Abas L., Benjamins R., Malenica N., Paciorek T., Wiśniewska J., Moulinier–Anzola J.C., Sieberer T., Friml J., Luschnig C. Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 2006;8:249–256. doi: 10.1038/ncb1369. [DOI] [PubMed] [Google Scholar]
- Adamowski M., Friml J. PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell. 2015;27:20–32. doi: 10.1105/tpc.114.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Barbosa I.C.R., Zourelidou M., Willige B.C., Weller B., Schwechheimer C. D6 PROTEIN KINASE activates auxin transport-dependent growth and PIN-FORMED phosphorylation at the plasma membrane. Dev. Cell. 2014;29:674–685. doi: 10.1016/j.devcel.2014.05.006. [DOI] [PubMed] [Google Scholar]
- Benjamins R., Scheres B. Auxin: the looping star in plant development. Annu. Rev. Plant Biol. 2008;59:443–465. doi: 10.1146/annurev.arplant.58.032806.103805. [DOI] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- Bennett T., Brockington S.F., Rothfels C., Graham S.W., Stevenson D., Kutchan T., Rolf M., Thomas P., Wong G.K.-S., Leyser O. Paralogous radiations of pin proteins with multiple origins of noncanonical pin structure. Mol. Biol. Evol. 2014;31:2042–2060. doi: 10.1093/molbev/msu147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleth T., Scarpella E., Prusinkiewicz P. Towards the systems biology of auxin-transport-mediated patterning. Trends Plant Sci. 2007;12:151–159. doi: 10.1016/j.tplants.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Bernsel A., Viklund H., Hennerdal A., Elofsson A. TOPCONS: consensus prediction of membrane protein topology. Nucl. Acids Res. 2009;37:W465–W468. doi: 10.1093/nar/gkp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433:39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- Bliss C.L., Novy F.G. Action of formaldehyde on enzymes and on certain pboteids. J. Exp. Med. 1899;4:47–80. doi: 10.1084/jem.4.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco C.D., Dovzhenko A., Liu X., Woerner N., Rensch T., Eismann M., Eimer S., Hegermann J., Paponov I.A., Ruperti B. The endoplasmic reticulum localized PIN8 is a pollen specific auxin carrier involved in intracellular auxin homeostasis. Plant J. 2012;71:860–870. doi: 10.1111/j.1365-313X.2012.05037.x. [DOI] [PubMed] [Google Scholar]
- Bowie J.U. Flip-flopping membrane proteins. Nat. Struct. Mol. Biol. 2006;13:94–96. doi: 10.1038/nsmb0206-94. [DOI] [PubMed] [Google Scholar]
- Brach T., Soyk S., Müller C., Hinz G., Hell R., Brandizzi F., Meyer A.J. Non-invasive topology analysis of membrane proteins in the secretory pathway. Plant J. 2009;57:534–541. doi: 10.1111/j.1365-313X.2008.03704.x. [DOI] [PubMed] [Google Scholar]
- Daley D.O., Rapp M., Granseth E., Melén K., Drew D., von Heijne G. Global topology analysis of the Escherichia coli inner membrane proteome. Science. 2005;308:1321–1323. doi: 10.1126/science.1109730. [DOI] [PubMed] [Google Scholar]
- Ding Z., Wang B., Moreno I., Dupláková N., Simon S., Carraro N., Reemmer J., Pěnčík A., Chen X., Tejos R. ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat. Commun. 2012;3:941. doi: 10.1038/ncomms1941. [DOI] [PubMed] [Google Scholar]
- Domingo B., Gasset M., Durán-Prado M., Castaño J., Serrano A., Fischer T., Llopis J. Discrimination between alternate membrane protein topologies in living cells using GFP/YFP tagging and pH exchange. Cell Mol. Life Sci. 2010;67:3345–3354. doi: 10.1007/s00018-010-0386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew D., Sjöstrand D., Nilsson J., Urbig T., Chin C., de Gier J.-W., von Heijne G. Rapid topology mapping of Escherichia coli inner-membrane proteins by prediction and PhoA/GFP fusion analysis. Proc. Natl. Acad. Sci. USA. 2002;99:2690–2695. doi: 10.1073/pnas.052018199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elofsson A., von Heijne G. Membrane protein structure: prediction versus reality. Annu. Rev. Biochem. 2007;76:125–140. doi: 10.1146/annurev.biochem.76.052705.163539. [DOI] [PubMed] [Google Scholar]
- Feraru E., Feraru M.I., Kleine-Vehn J., Martinière A., Mouille G., Vanneste S., Vernhettes S., Runions J., Friml J. Pin polarity maintenance by the cell wall in Arabidopsis. Curr. Biol. 2011;21:338–343. doi: 10.1016/j.cub.2011.01.036. [DOI] [PubMed] [Google Scholar]
- Friml J., Benková E., Mayer U., Palme K., Muster G. Automated whole mount localisation techniques for plant seedlings. Plant J. 2003;34:115–124. doi: 10.1046/j.1365-313x.2003.01705.x. [DOI] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- Friml J., Yang X., Michniewicz M., Weijers D., Quint A., Tietz O., Benjamins R., Ouwerkerk P.B.F., Ljung K., Sandberg G. A PINOID-dependent binary switch in apical-basal pin polar targeting directs auxin efflux. Science. 2004;306:862–865. doi: 10.1126/science.1100618. [DOI] [PubMed] [Google Scholar]
- Furutani M., Vernoux T., Traas J., Kato T., Tasaka M., Aida M. PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development. 2004;131:5021–5030. doi: 10.1242/dev.01388. [DOI] [PubMed] [Google Scholar]
- Gälweiler L., Guan C., Müller A., Wisman E., Mendgen K., Yephremov A., Palme K. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;282:2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- Ganguly A., Sasayama D., Cho H.-T. Regulation of the polarity of protein trafficking by phosphorylation. Mol. Cells. 2012;33:423–430. doi: 10.1007/s10059-012-0039-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A., Park M., Kesawat M.S., Cho H.-T. Functional analysis of the hydrophilic loop in intracellular trafficking of Arabidopsis PIN-FORMEDproteins. Plant Cell. 2014;26:1570–1585. doi: 10.1105/tpc.113.118422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjetting S.K., Ytting C.K., Schulz A., Fuglsang A.T. Live imaging of intra- and extracellular pH in plants using pHusion, a novel genetically encoded biosensor. J. Exp. Bot. 2012;63:3207–3218. doi: 10.1093/jxb/ers040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood D. Theoretical and practical aspects of glutaraldehyde fixation. Histochem. J. 1972;4:267–303. doi: 10.1007/BF01005005. [DOI] [PubMed] [Google Scholar]
- Huang F., Kemel Zago M., Abas L., van Marion A., Galván-Ampudia C.S., Offringa R. Phosphorylation of conserved pin motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell. 2010;22:1129–1142. doi: 10.1105/tpc.109.072678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.T. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics. 2007;23:538–544. doi: 10.1093/bioinformatics/btl677. [DOI] [PubMed] [Google Scholar]
- Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protocols. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Melén K., von Heijne G. Topology models for 37 Saccharomyces cerevisiae membrane proteins based on C-terminal reporter fusions and predictions. J. Biol. Chem. 2003;278:10208–10213. doi: 10.1074/jbc.M300163200. [DOI] [PubMed] [Google Scholar]
- Kitakura S., Vanneste S., Robert S., Löfke C., Teichmann T., Tanaka H., Friml J. Clathrin mediates endocytosis and polar distribution of pin auxin transporters in Arabidopsis. Plant Cell. 2011;23:1920–1931. doi: 10.1105/tpc.111.083030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Dhonukshe P., Sauer M., Brewer P.B., Wiśniewska J., Paciorek T., Benková E., Friml J. Arf GEF-dependent transcytosis and polar delivery of pin auxin carriers in Arabidopsis. Curr. Biol. 2008;18:526–531. doi: 10.1016/j.cub.2008.03.021. [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Leitner J., Zwiewka M., Sauer M., Abas L., Luschnig C., Friml J. Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. PNAS. 2008;105:17812–17817. doi: 10.1073/pnas.0808073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneen M., Farinas J., Li Y., Verkman A.S. Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys. J. 1998;74:1591–1599. doi: 10.1016/S0006-3495(98)77870-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Křeček P., Skůpa P., Libus J., Naramoto S., Tejos R., Friml J., Zažímalová E. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 2009;10:249. doi: 10.1186/gb-2009-10-12-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K. Auxin metabolism and homeostasis during plant. Development. 2013;140:943–950. doi: 10.1242/dev.086363. [DOI] [PubMed] [Google Scholar]
- Llopis J., McCaffery J.M., Miyawaki A., Farquhar M.G., Tsien R.Y. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc. Natl. Acad. Sci. USA. 1998;95:6803–6808. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz H., Hailey D.W., Lippincott-Schwartz J. Fluorescence protease protection of GFP chimeras to reveal protein topology and subcellular localization. Nat. Meth. 2006;3:205–210. doi: 10.1038/nmeth857. [DOI] [PubMed] [Google Scholar]
- Mayor S., Riezman H. Sorting GPI-anchored proteins. Nat. Rev. Mol. Cell Biol. 2004;5:110–120. doi: 10.1038/nrm1309. [DOI] [PubMed] [Google Scholar]
- Michniewicz M., Zago M.K., Abas L., Weijers D., Schweighofer A., Meskiene I., Heisler M.G., Ohno C., Zhang J., Huang F. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell. 2007;130:1044–1056. doi: 10.1016/j.cell.2007.07.033. [DOI] [PubMed] [Google Scholar]
- Migneault I., Dartiguenave C., Bertrand M.J., Waldron K.C. Glutaraldehyde: behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. BioTechniques. 2004;37:790–796. doi: 10.2144/04375RV01. 798–802. [DOI] [PubMed] [Google Scholar]
- Movafeghi A., Happel N., Pimpl P., Tai G.-H., Robinson D.G. Arabidopsis Sec21p and Sec23p homologs. Probable coat proteins of plant COP-coated vesicles. Plant Physiol. 1999;119:1437–1446. doi: 10.1104/pp.119.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec J., Skůpa P., Bailly A., Hoyerová K., Krecek P., Bielach A., Petrásek J., Zhang J., Gaykova V., Stierhof Y.-D. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature. 2009;459:1136–1140. doi: 10.1038/nature08066. [DOI] [PubMed] [Google Scholar]
- Nilsson J., Persson B., von Heijne G. Consensus predictions of membrane protein topology. FEBS Lett. 2000;486:267–269. doi: 10.1016/s0014-5793(00)02321-8. [DOI] [PubMed] [Google Scholar]
- Nilsson J., Persson B., von Heijne G. Prediction of partial membrane protein topologies using a consensus approach. Protein Sci. 2002;11:2974–2980. doi: 10.1110/ps.0226702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisar N., Cuttriss A.J., Pogson B.J., Cazzonelli C.I. The promoter of the Arabidopsis PIN6 auxin transporter enabled strong expression in the vasculature of roots, leaves, floral stems and reproductive organs. Plant Signal Behav. 2014;9:e27898. doi: 10.4161/psb.27898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad N., Shichrur K., Yalovsky S. The analysis of protein-protein interactions in plants by bimolecular fluorescence complementation. Plant Physiol. 2007;145:1090–1099. doi: 10.1104/pp.107.107284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Ueda J., Komaki M.K., Bell C.J., Shimura Y. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell. 1991;3:677–684. doi: 10.1105/tpc.3.7.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterrieder A., Carvalho C.M., Latijnhouwers M., Johansen J.N., Stubbs C., Botchway S., Hawes C. Fluorescence lifetime imaging of interactions between Golgi tethering factors and small GTPases in plants. Traffic. 2009;10:1034–1046. doi: 10.1111/j.1600-0854.2009.00930.x. [DOI] [PubMed] [Google Scholar]
- Paciorek T., Zazímalová E., Ruthardt N., Petrásek J., Stierhof Y.-D., Kleine-Vehn J., Morris D.A., Emans N., Jürgens G., Geldner N. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature. 2005;435:1251–1256. doi: 10.1038/nature03633. [DOI] [PubMed] [Google Scholar]
- Palme K., Gälweiler L. PIN-pointing the molecular basis of auxin transport. Curr. Opin. Plant Biol. 1999;2:375–381. doi: 10.1016/s1369-5266(99)00008-4. [DOI] [PubMed] [Google Scholar]
- Petrášek J., Mravec J., Bouchard R., Blakeslee J.J., Abas M., Seifertová D., Wiśniewska J., Tadele Z., Kubeš M., Čovanová M. Pin proteins perform a rate-limiting function in cellular auxin efflux. Science. 2006;312:914–918. doi: 10.1126/science.1123542. [DOI] [PubMed] [Google Scholar]
- Pimpl P., Movafeghi A., Coughlan S., Denecke J., Hillmer S., Robinson D.G. In situ localization and in vitro induction of plant COPI-coated vesicles. Plant Cell. 2000;12:2219–2235. doi: 10.1105/tpc.12.11.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogorelko G., Fursova O., Lin M., Pyle E., Jass J., Zabotina O. Post-synthetic modification of plant cell walls by expression of microbial hydrolases in the apoplast. Plant Mol. Biol. 2011;77:433–445. doi: 10.1007/s11103-011-9822-9. [DOI] [PubMed] [Google Scholar]
- Rapp M., Drew D., Daley D.O., Nilsson J., Carvalho T., Melén K., De Gier J.-W., von Heijne G. Experimentally based topology models for E. coli inner membrane proteins. Protein Sci. 2004;13:937–945. doi: 10.1110/ps.03553804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp M., Granseth E., Seppälä S., von Heijne G. Identification and evolution of dual-topology membrane proteins. Nat. Struct. Mol. Biol. 2006;13:112–116. doi: 10.1038/nsmb1057. [DOI] [PubMed] [Google Scholar]
- Robert S., Chary S.N., Drakakaki G., Li S., Yang Z., Raikhel N.V., Hicks G.R. Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. Proc. Natl. Acad. Sci. USA. 2008;105:8464–8469. doi: 10.1073/pnas.0711650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert H.S., Crhak Khaitova L., Mroue S., Benková E. The importance of localized auxin production for morphogenesis of reproductive organs and embryos in Arabidopsis. J. Exp. Bot. 2015;66:5029–5042. doi: 10.1093/jxb/erv256. [DOI] [PubMed] [Google Scholar]
- Russell A.D., Hopwood D. The biological uses and importance of glutaraldehyde. Prog. Med. Chem. 1976;13:271–301. doi: 10.1016/s0079-6468(08)70140-1. [DOI] [PubMed] [Google Scholar]
- Sabatini D.D., Bensch K., Barrnett R.J. Cytochemistry and electron microscopy. J. Cell Biol. 1963;17:19–58. doi: 10.1083/jcb.17.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M., Paciorek T., Benková E., Friml J. Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat. Protoc. 2006;1:98–103. doi: 10.1038/nprot.2006.15. [DOI] [PubMed] [Google Scholar]
- Schwacke R., Schneider A., Van Der Graaff E., Fischer K., Catoni E., Desimone M., Frommer W.B., Flügge U.-I., Kunze R. ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 2003;131:16–26. doi: 10.1104/pp.011577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T. Protein modeling: what happened to the “protein structure gap”? Structure. 2013;21:1531–1540. doi: 10.1016/j.str.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook J.C., Carroll K.L., Hung K.F., Masson P.H., Somerville C.R. The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell. 2002;14:1635–1648. doi: 10.1105/tpc.002360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S., Petrášek J. Why plants need more than one type of auxin. Plant Sci. 2011;180:454–460. doi: 10.1016/j.plantsci.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Sparkes I., Tolley N., Aller I., Svozil J., Osterrieder A., Botchway S., Mueller C., Frigerio L., Hawes C. Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell. 2010;22:1333–1343. doi: 10.1105/tpc.110.074385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., Friml J., Marchant A., Ljung K., Sandberg G., Palme K., Bennett M. Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 2001;15:2648–2653. doi: 10.1101/gad.210501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., Kargul J., Marchant A., Zadik D., Rahman A., Mills R., Yemm A., May S., Williams L., Millner P. Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell. 2004;16:3069–3083. doi: 10.1105/tpc.104.024737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K., Benková E., Swarup R., Casimiro I., Péret B., Yang Y., Parry G., Nielsen E., De Smet I., Vanneste S. The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 2008;10:946–954. doi: 10.1038/ncb1754. [DOI] [PubMed] [Google Scholar]
- Tanaka K.A.K., Suzuki K.G.N., Shirai Y.M., Shibutani S.T., Miyahara M.S.H., Tsuboi H., Yahara M., Yoshimura A., Mayor S., Fujiwara T.K. Membrane molecules mobile even after chemical fixation. Nat. Meth. 2010;7:865–866. doi: 10.1038/nmeth.f.314. [DOI] [PubMed] [Google Scholar]
- Vanneste S., Friml J. Auxin: a trigger for change in plant development. Cell. 2009;136:1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Viaene T., Delwiche C.F., Rensing S.A., Friml J. Origin and evolution of PIN auxin transporters in the green lineage. Trends Plant Sci. 2013;18:5–10. doi: 10.1016/j.tplants.2012.08.009. [DOI] [PubMed] [Google Scholar]
- Vieten A., Vanneste S., Wiśniewska J., Benková E., Benjamins R., Beeckman T., Luschnig C., Friml J. Functional redundancy of pin proteins is accompanied by auxin-dependent cross-regulation of pin expression. Development. 2005;132:4521–4531. doi: 10.1242/dev.02027. [DOI] [PubMed] [Google Scholar]
- Vieten A., Sauer M., Brewer P.B., Friml J. Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci. 2007;12:160–168. doi: 10.1016/j.tplants.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Webster J.D., Miller M.A., DuSold D., Ramos-Vara J. Effects of prolonged formalin fixation on diagnostic immunohistochemistry in domestic animals. J. Histochem. Cytochem. 2009;57:753–761. doi: 10.1369/jhc.2009.953877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewska J., Xu J., Seifertová D., Brewer P.B., Růžička K., Blilou I., Rouquié D., Benková E., Scheres B., Friml J. Polar PIN localization directs auxin flow in plants. Science. 2006;312:883. doi: 10.1126/science.1121356. [DOI] [PubMed] [Google Scholar]
- Xi W., Gong X., Yang Q., Yu H., Liou Y.-C. Pin1At regulates PIN1 polar localization and root gravitropism. Nat. Commun. 2016;7:10430. doi: 10.1038/ncomms10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Scheres B. Dissection of Arabidopsis ADP-RIBOSYLATIONFACTOR 1 function in epidermal cell polarity. Plant Cell. 2005;17:525–536. doi: 10.1105/tpc.104.028449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Hofhuis H., Heidstra R., Sauer M., Friml J., Scheres B. A molecular framework for plant regeneration. Science. 2006;311:385–388. doi: 10.1126/science.1121790. [DOI] [PubMed] [Google Scholar]
- Yang H., Murphy A.S. Functional expression and characterization of Arabidopsis ABCB, AUX 1 and PIN auxin transporters in Schizosaccharomyces pombe. Plant J. 2009;59:179–191. doi: 10.1111/j.1365-313X.2009.03856.x. [DOI] [PubMed] [Google Scholar]
- Žádníková P., Petrášek J., Marhavý P., Raz V., Vandenbussche F., Ding Z., Schwarzerová K., Morita M.T., Tasaka M., Hejátko J. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development. 2010;137:607–617. doi: 10.1242/dev.041277. [DOI] [PubMed] [Google Scholar]
- Zhang J., Nodzyński T., Pěnčík A., Rolčík J., Friml J. PIN phosphorylation is sufficient to mediate pin polarity and direct auxin transport. PNAS. 2010;107:918–922. doi: 10.1073/pnas.0909460107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zourelidou M., Absmanner B., Weller B., Barbosa I.C., Willige B.C., Fastner A., Streit V., Port S.A., Colcombet J., de la Fuente van Bentem S. Auxin efflux by PIN-FORMED proteins is activated by two different protein kinases, D6 PROTEIN KINASE and PINOID. Elife. 2014;3:e02860. doi: 10.7554/eLife.02860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.