Summary
Compartmentalization of the gastrointestinal (GI) tract of metazoans is critical for health. GI compartments contain specific microbiota, and microbiota dysbiosis is associated with intestinal dysfunction. Dysbiosis develops in aging intestines, yet how this relates to changes in GI compartmentalization remains unclear. The Drosophila GI tract is an accessible model to address this question. Here we show that the stomach-like copper cell region (CCR) in the middle midgut controls distribution and composition of the microbiota. We find that chronic activation of JAK/Stat signaling in the aging gut induces a metaplasia of the gastric epithelium, CCR decline, and subsequent commensal dysbiosis and epithelial dysplasia along the GI tract. Accordingly, inhibition of JAK/Stat signaling in the CCR specifically prevents age-related metaplasia, commensal dysbiosis and functional decline in old guts, and extends lifespan. Our results establish a mechanism by which age-related chronic inflammation causes the decline of intestinal compartmentalization and microbiota dysbiosis, limiting lifespan.
Introduction
The intestinal microbiota influences long-term homeostasis of metazoans by maintaining epithelial integrity in the GI tract, supporting digestion, training the intestinal immune system, and preventing the growth of pathogenic bacteria (Clemente et al., 2012). The composition of the gut microbiota is influenced by the interaction with GI epithelia and by a range of genetic and environmental factors (David et al., 2014; Clemente et al., 2012). Alteration of microbiota composition (‘dysbiosis’) has been associated with numerous pathologies and with aging (Clemente et al., 2012; Claesson et al., 2011; Claesson et al., 2012). While preventing commensal dysbiosis in aging flies extends lifespan, its origin is unknown (Buchon et al., 2013; Clark et al., 2015; Guo et al., 2014).
The GI tract of most animals is lined by a series of highly specialized epithelia that perform localized functions, but have common characteristics to manage the commensal population, allow immune responses to infections, and maintain intestinal barrier function (Barker et al., 2010; Buchon et al., 2013; Li et al., 2013; Marianes and Spradling, 2013; Strand and Micchelli, 2011). How compartmentalization influences the microbiota, and whether age-related changes in compartment identities contribute to age-related dysbiosis, remains unknown.
The adult Drosophila intestine constitutes a genetically accessible model system to address these questions. Studies exploring age-related changes in epithelial homeostasis, regenerative capacity, stem cell (SC) function, host-commensal interactions, and innate immune signaling have provided rich insight into mechanisms governing intestinal homeostasis (Ayyaz and Jasper, 2013; Biteau et al., 2011; Lemaitre and Miguel-Aliaga, 2013). Based on morphological and functional characteristics, the midgut of flies can be subdivided roughly into the anterior midgut (AM), the middle midgut (MM), which contains an acidic gastric or copper cell region (CCR (Dubreuil, 2004)), and the posterior midgut (PM; Fig. 1A). Finer subdivisions into 10–14 regions have been described based on more detailed morphological and molecular landmarks along the GI tract (Buchon et al., 2013; Marianes and Spradling, 2013). Intestinal SCs (ISCs) can be found in each of these compartments (Biteau et al., 2011; Strand and Micchelli, 2011). ISCs in the PM express escargot (esg) and Delta (Dl), and divide asymmetrically to give rise to precursor cells (the Dl−/esg+ Enteroblasts, EBs), which will further differentiate into either Pdm1 - expressing Enterocytes (ECs) or prospero (pros) - expressing Enteroendocrine cells (EEs) (Ayyaz and Jasper, 2013; Biteau et al., 2011; Lemaitre and Miguel-Aliaga, 2013). In the CCR, esg+ gastric stem cells (GSSCs) respond to stress by inducing regeneration of three different cell types: Dve+/Labial+/Cut+ Copper cells (CCs, which secrete hydrochloric acid), Dve+/weak Labial+/Cut-interstitial cells, and Pros+ EEs (Strand and Micchelli, 2011) (Fig. S2A). A gradient of the morphogen Decapentaplegic (Dpp), a Drosophila Bmp2/4 orthologue, segregates GSSCs from posterior midgut ISCs (Li et al., 2013).
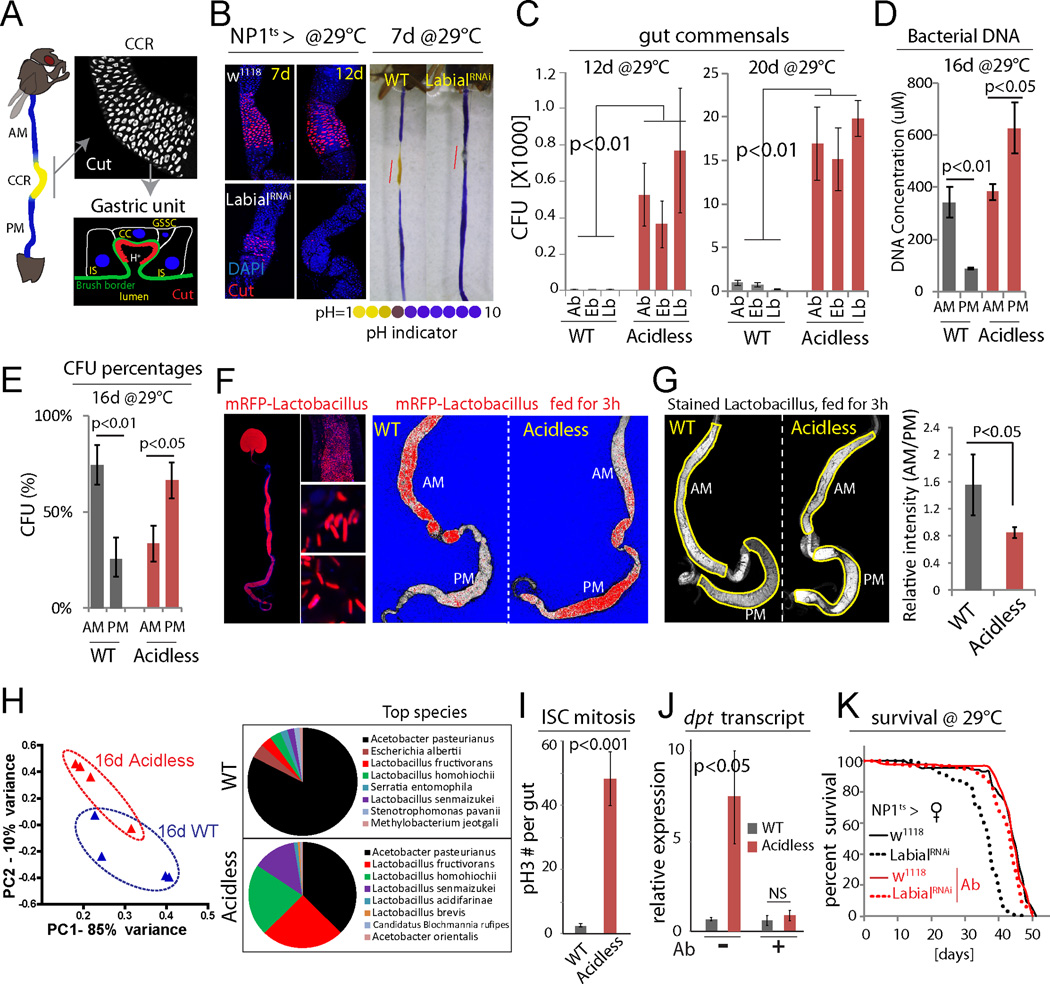
Figure 1. Intestinal compartmentalization impacts gut microbiota.
(A) Left: the Drosophila GI tract consists of anterior midgut (AM), middle midgut (MM) with copper cell region (CCR), and posterior midgut (PM). Right: anti-Cut (white) labels acid-producing copper cells (CCs). A gastric unit. IS: interstitial cell, GSSC: gastric stem cell.
(B) Expression of LabialRNAi results in loss of CCs. Acidic regions (yellow) indicated by pH indicator Bromophenol blue. Representative images of 3 independent experiments. N=7 for each genotype and time point.
(C) Commensal numbers in 12d and 20d old WT (NP1ts>w1118) and acidless flies (NP1ts>LabialRNAi). Colony-forming units (CFUs) of three common commensal phylotypes (Acetobacter, Ab; Enterobacter, Eb; Lactobacillus, Lb) are quantified using selective plates. Averages and s.e.m. (t-test), N=6, 6, 6, 10, 10, 10 (both 12d and 20d). Representative from 3 independent experiments.
(D) Bacterial DNA concentration of the AM and PM from 16d old WT and acidless flies. Averages and s.e.m. (t-test), N=3×10 guts each.
(E) Commensal distribution in 16d old WT and acidless guts. Percent averages of colony-forming units (CFUs) in AMs and PMs are shown. N=12, 11. Averages and s.e.m. (t-test).
(F) Distribution of RFP-tagged Lactobacillus plantarum in 16d old WT and acidless flies. Left: representative images of RFP-tagged Lactobacillus in the gut. Right: representative saturation images of WT and acidless guts with RFP-tagged Lactobacillus. Saturated signal red, un-saturated signal white and background blue. False color images generated from confocal images in ZEN. Representative of 3 independent experiments.
(G) Distribution of stained Lactobacillus (by SYTO 9) in 16d old WT and acidless flies. Relative fluorescent intensity is quantified. N=6, 9. Averages and s.e.m. (t-test).
(H) Commensal composition of 16d old WT and acidless guts. Principal component analysis (PCA) based on percentages of classified bacterial species determined by 16S rRNA sequencing. Pie charts show top bacterial species. For PCA, each dot represents 10 guts; for pie charts, each represents the average of four samples (4×10 guts).
(I) Quantification of pH3+ cells in guts of 16d old flies at 29°C. Averages and s.e.m. (t-test); N=12, 11. Data are representative of 3 independent experiments.
(J) qRT-PCR showing dpt expression of 16d old flies on normal or antibiotic (Ab) food. Averages and s.e.m. (t-test) of N=3 biological replicates.
(K) Lifespan of acidless flies compared to WT controls on normal or antibiotic (Ab) food. Without antibiotics: WT N=88, acidless N=93, Log-rank P value <0.0001, 17.8% change of median lifespan; Antibiotics: WT N=110, acidless N=100, Log-rank P value =0.02, no change of median lifespan.
See also Fig. S1
The aging Drosophila intestine develops an inflammatory condition that is characterized by immune dysfunction, commensal dysbiosis and intestinal stem cell (ISC) over-proliferation (Biteau et al., 2010a; Buchon et al., 2009; Clark et al., 2015; Guo et al., 2014). This results in metabolic decline and is associated with loss of the intestinal barrier function, promoting mortality in aging animals (Ayyaz and Jasper, 2013; Biteau et al., 2011; Biteau et al., 2010b; Guo et al., 2014; Lemaitre and Miguel-Aliaga, 2013; Rera et al., 2012). Interventions that limit dysbiosis and ISC over-proliferation extend lifespan (Biteau et al., 2010b; Clark et al., 2015; Guo et al., 2014), yet the underlying cause for the age-related loss of intestinal homeostasis remains unclear. Previous studies have found that intestinal compartmentalization declines with age (Buchon et al., 2013) and that loss of Caudal, a transcription factor specifically expressed in the posterior midgut, impacts immune homeostasis and commensal composition (Ryu et al., 2008). Whether and how GI compartmentalization impacts the microbiota, whether changes in GI compartmentalization are associated with aging, and whether such changes influences longevity in flies has not been tested.
Results
Gut compartmentalization and microbiota
To characterize the importance of GI compartmentalization for the maintenance and composition of the gut microbiota, we generated flies in which the stomach-like CCR was specifically ablated by knocking down Labial using NP1::Gal4, tub::Gal80ts, a temperature sensitive driver that is specific for differentiated cells in the gut ((McGuire et al., 2004a); this combination is referred to as NP1ts; Fig. 1B, S1A, B). Since Labial is required for CC maintenance (Buchon et al., 2013; Li et al., 2013), this results in rapid decline of CCs, and a corresponding loss of acidity in the MM. In these ‘acidless’ flies, the expression of the CC-specific vacuolar-type H+ ATPase, Vha100-4, was significantly reduced compared to controls, confirming the loss of CCs (Fig. S1D, E).
In acidless flies, the number of colony forming units (CFUs) of the three main commensal phylotypes (Lactobacilli, Acetobacilli and Enterobacteria) increased significantly (Fig. 1C; Fig. S1C, F, G). Since commensals in flies are only weakly associated with the gut and need continuous reconstitution by ingestion (Blum et al., 2013; Broderick et al., 2014), we hypothesized that the CCR manages survival of ingested bacteria and colonization of the PM. Supporting this hypothesis, the bacterial load (the number of CFUs, or the concentration of bacterial DNA) is higher in the AM than the PM of wild-type flies, but this relationship is reversed in acidless flies (Fig. 1D, E, S1H; note that there is also an increase in commensals in the AM in acidless animals, potentially due to re-ingestion of excreted bacteria, or due to immune dysfunction triggered by elevated Foxo activity in ECs of the AM, see below). Furthermore, the distribution of RFP-tagged commensal Lactobacillus plantarum isolated from wild-type flies and reintroduced into wild-type or acidless flies is strongly impacted by the CCR: tagged bacteria are enriched in the AM of wild-type flies, but are predominantly localized in the PM of acidless flies (Fig. 1F, S1I). The same was observed using Lactobacilli stained by a fluorescent dye (Fig. 1G).
16S rRNA gene sequencing revealed a significant difference in microbiota composition between wild-type and acidless guts, reflecting a strong enrichment of Lactobacilli at the expense of Acetobacteria (Fig. 1H). Acidless flies further exhibit increased expression of the Foxo target gene thor (Fig. S1J, K; note that the most obvious increase was seen in the AM and the posterior PM) and of the anti-microbial peptide Diptericin (Dpt), as well as higher rates of ISC proliferation compared to wild-type (as determined by quantifying mitotic figures, phospho-Histone H3 expressing cells, Fig.1I, J, S1L). These phenotypes are reminiscent of the loss of intestinal homeostasis in aging animals, which is characterized by elevated Foxo activity in ECs, resulting in hyperactivation of the Relish/NFkB signaling pathway (which induces Dpt), commensal dysbiosis, and ISC hyperactivation (Guo et al., 2014). Accordingly, acidless flies exhibit shorter lifespan than controls (Fig. 1K). Antibiotic treatment is sufficient to reduce Dpt expression and restores wild-type lifespan in acidless animals (Fig. 1J, K), confirming that commensal dysbiosis triggers these phenotypes.
Age-related gastric decline and metaplasia
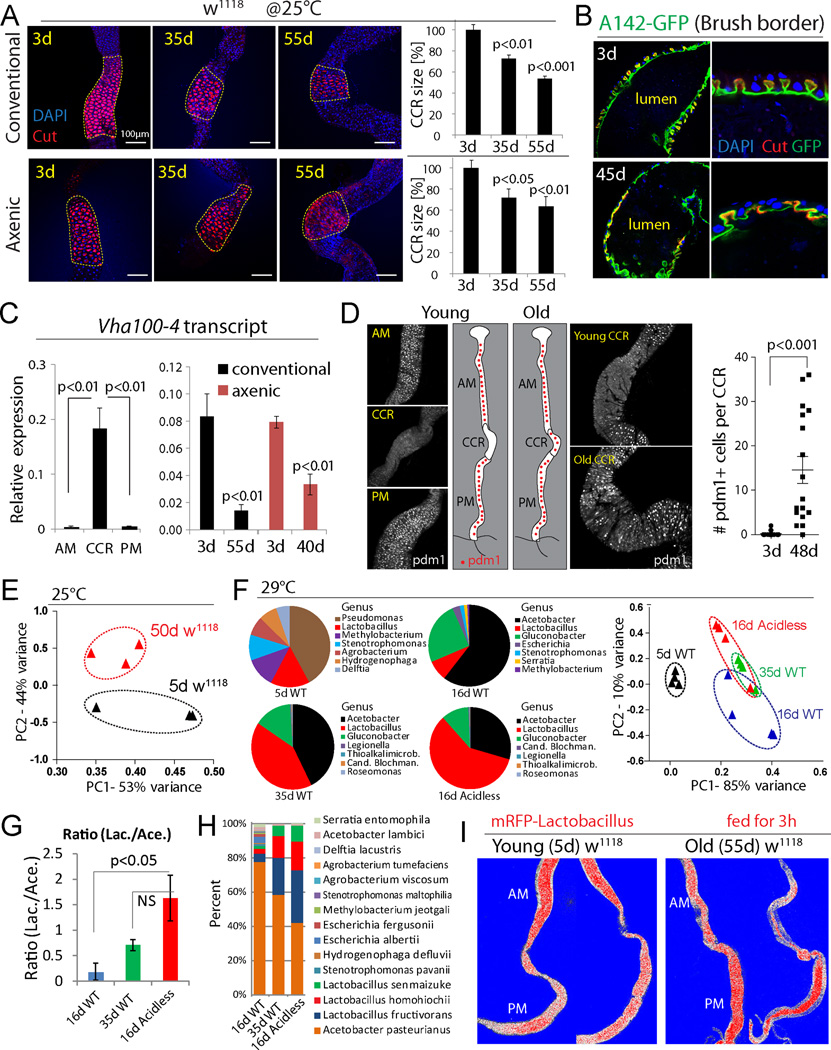
Since these results uncovered a critical role for the CCR in managing the microbiota, and since acidless flies exhibited phenotypes reminiscent of old flies, we assessed CCR function in aging wild-type flies. We find that the size of the CCR and the structure of gastric units decline with age in both conventionally and axenically reared animals (Fig. 2A, B). This is associated with reduced expression of Vha100-4 (Fig. 2C, S2B), and with accumulation of Pdm1+ cells in the CCR (Fig. 2D, S2C, D).
Figure 2. Age-related gastric decline and metaplasia.
(A) CCRs (outlined) of w1118 flies decline with age. CCR area is quantified on the right. Averages and s.e.m. (t-test) from one experiment representative of more than three independent repeats. Conventional: N=5, 7, 7; Axenic: N=7, 11, 11.
(B) Age-related decline of gastric units of w1118 flies. Anti-Cut (red) and brush border (green, A142-GFP). Representative images of 6 flies at each age in 2 independent experiments.
(C) qRT-PCR measuring expression of Vha100-4 (normalized to actin5C) in different regions of young w1118 flies (left) and in whole guts of w1118 flies (right). Averages and s.e.m. (t-test), N=3 biological replicates.
(D) Pdm1 expression along the GI tract. Representative images, experiment has been repeated at least three times in three wild-type genetic backgrounds (y1,w1/w1118; Dve::Gal4/+, y1,w1/w1118; w1118, S1106::GS/+). Pdm1+ cells per CCR are quantified. Averages and s.e.m. (t-test). N=20, 17.
(E) Principal component analysis (PCA) based on percentages of classified bacterial species from 5d and 50d old w1118 flies at 25°C. Each dot represents 10 guts.
(F) Pie charts show top bacterial genera of 5d old WT (NP1ts>w1118), 16d WT, 35d WT and 16d acidless (NP1ts>LabialRNAi) flies. Flies are cultured at 29°C to induce driver expression. PCA is based on percentages of classified bacterial species. For PCA, each dot represents 10 guts; for pie charts, each represents the average of four samples (40 guts).
(G, H) Ratios of total Lactobacillus and Acetobacter (G), and distribution of top 15 bacterial species (H), in flies of three conditions shown in (F). Averages and s.e.m. (t-test) for (G). Averages of four samples for (H).
(I) Change of Lactobacillus distribution in old guts (compare Fig. 1F).
See also Fig. S2
Vha100-4 is a member of a family of vacuolar-type H+ ATPases encoded in the fly genome. Vha16-1 has been reported to be expressed in the CCR based on a Gal4 trap line, and to be required to maintain gut acidity (Lin et al, 2015). However, our RT-PCR and RNAseq data indicate that Vha100-4, which is highly enriched in the CCR (Fig. 2C), is the only Vha gene with decreased expression in guts of acidless flies (Fig. S1D). Another study (Buchon, 2013) has found high specific Vha100-4 expression in the CCR, while Vha16-1 cannot be detected. Systematic analysis of Vha genes and their relative functions in the fly gut is of interest for future studies.
Pdm1 (also called nubbin), a POU-domain homeobox transcription factor, is a widely used marker for ECs in the PM (Lee et al., 2009) (Fig. S2D), and is required for their differentiation (Korzelius et al., 2014), but is not expressed in cells of the MM in young flies (Fig. 2D, S2D; no change in Pdm1 expression is seen in the aging PM, Fig. S2E). The appearance of ectopic Pdm1+ cells suggested that the age-related decline of the CCR is the result of a metaplasia (a condition in which epithelial subtypes acquire ectopic properties) in which CCs in the MM are replaced by Pdm1+ EC-like cells characteristic of the PM epithelium.
Since this gastric decline occurs in both axenic and conventional conditions, it is likely a cause, not a consequence of the age-related loss of intestinal homeostasis. Supporting this notion, the microbiota composition of young acidless animals resembles that of old wild-type flies more than that of wild-type flies of the same age (Fig. 2E–H, S2F, G; the composition in old wild-type flies differs significantly from that of young wild-type flies reared at the same temperature). Furthermore, the anterior-posterior distribution of RFP-tagged Lactobacillus in old flies shows a pattern similar to that of acidless flies: tagged bacteria are enriched in the PM (Fig. 1F, 2I).
JAK/Stat activation causes CCR metaplasia
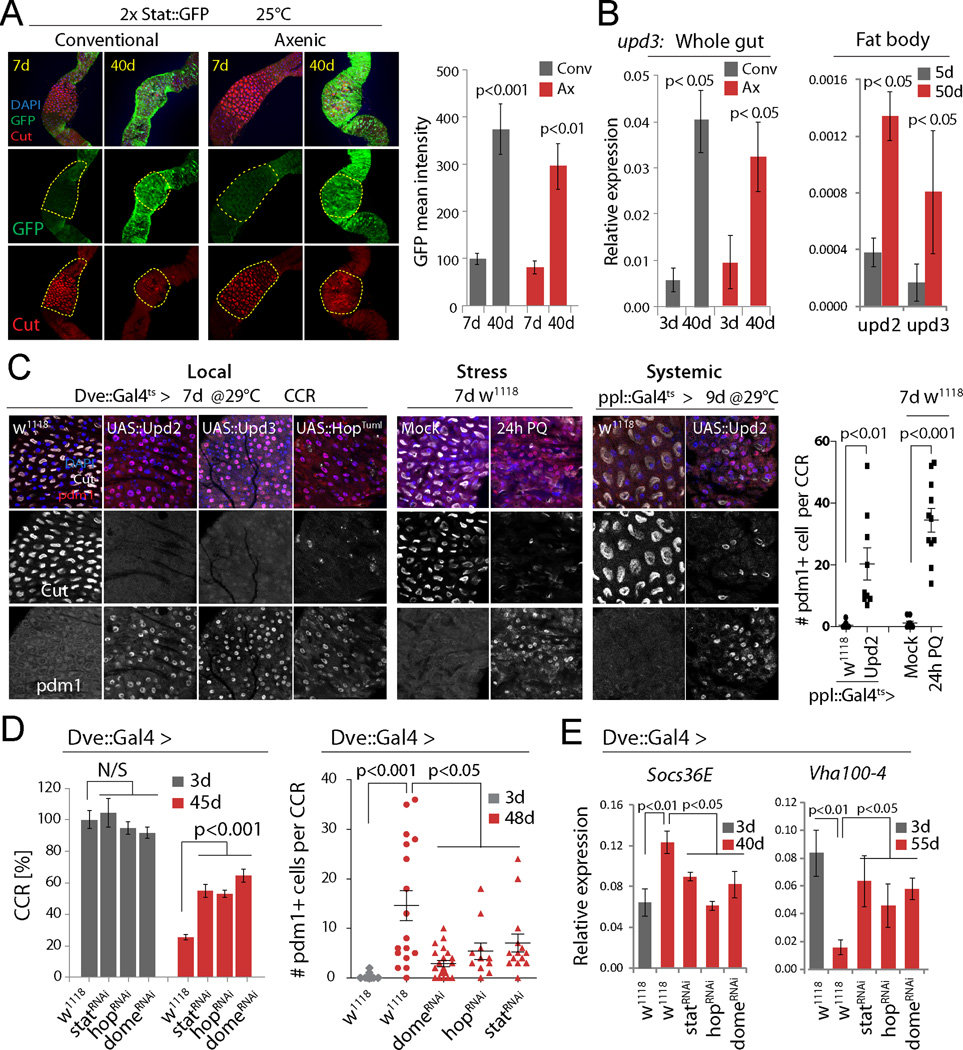
Age-related metaplasia thus significantly perturbs commensal distribution, composition, and size, and contributes to age-related intestinal dysfunction. To characterize the molecular mechanism causing this metaplasia, we explored signaling pathways whose activity changes with age in the gut of both axenically and conventionally reared animals (Guo et al., 2014). These pathways include JAK/Stat signaling, as illustrated by the fact that the expression of the JAK/Stat activity reporter 2xStat::GFP (Bach et al., 2007) increases throughout the gut (including the CCR) in flies aged under both conventional and axenic conditions (Fig. 3A). This activation is likely a result of local and/or systemic induction of one of the three Unpaired ligands (Upd, Upd2 and Upd3) that activate JAK/Stat signaling in flies. Indeed, expression of unpaired 3 (upd3) strongly increases with age in the gut, while upd2 and upd3 are significantly induced in the abdominal fatbody with age (Fig. 3B, S3A).
Figure 3. Age-related activation of JAK/Stat causes CC loss and metaplasia.
(A) Expression of 2xStat::GFP (green) in young and old CCRs. GFP intensity quantified on the right (normalized to 100 for young conv. condition). Averages and s.e.m. (t-test). Conv.: N=8, 8. Axenic: N=5, 9. Representative of 3 independent experiments.
(B) qRT-PCR measuring upd3 expression (normalized to actin5C) in guts, and upd2 and upd3 in fat bodies, of w1118 flies. Averages and s.e.m. (t-test). N=3 biological replicates.
(C) Immunostaining detecting Cut+ CCs and Pdm1+ cells in the CCR. (Local) wild-type flies or flies over-expressing Upd2, Upd3, or HopTuml. (Stress) treatment with 80mM Paraquat (PQ). (Systemic) over-expression of Upd2 using the fat body driver ppl::Gal4ts. Quantifications of Pdm1+ cells per CCR from stress and systemic conditions are shown. Averages and s.e.m. (t-test), N=12, 10, 10, 12. Data are representative of 3 independent experiments for Local, and 2 independent experiments for Stress and Systemic conditions.
(D) Knockdown of JAK/Stat signaling components limits age-induced gastric decline. Left: N=12, 11, 11, 12, 20, 20, 16, 22. Right: N=20, 17, 22, 12, 15. Averages and s.e.m. (t-test). Representative from 3 independent experiments.
(E) qRT-PCR detecting Socs36E and Vha100-4 in guts of wild-type (w1118) or StatRNAi, HopRNAi or DomeRNAi expressing flies. Averages and s.e.m. (t-test). N=3 biological replicates.
See also Fig. S3
Activation of JAK/Stat signaling in CCs of young guts by locally expressing Upd2 or Upd3 (using Dve::Gal4, which drives expression in differentiated cells of the MM, combined with tub::Gal80ts; Fig. S3B), by cell-autonomously activating JAK (using Dvets to express the constitutively active JAK/hopscotch mutant HopTumL, (Classen et al., 2009)), or by systemically elevating Upd2 expression (using the fatbody driver ppl::Gal4ts), resulted in strong metaplasia, widely replacing Cut+ cells by Pdm1+ cells in the MM (Fig. 3C; pplts-mediated expression of Upd3 did not impact the gut, not shown). Upd2 expression from fatbody is sufficient to activate JAK/Stat in the gut, as determined by the induction of the JAK/Stat target gene Socs36E (Fig. S3D). Metaplasia was also observed when JAK/Stat was activated acutely in the intestine of young flies by exposure to Paraquat, which damages ECs and triggers regenerative responses by broadly activating JNK and JAK/Stat signaling in the gut (including in the visceral muscle, EBs, and ISCs; Fig. 3C, S3C)(Biteau et al., 2008; Jiang et al., 2009; Osman et al., 2012; Zhou et al., 2013).
These data suggested that in old flies, both systemically and locally derived Upd ligands may contribute to constitutively activating JAK/Stat signaling in the gut, resulting in metaplasia of the CCR. Supporting this notion, knockdown of Stat, Hop or the receptor Dome in MM cells limits age-related or Paraquat-induced metaplasia (Fig. 3D, S3C, G, H), reduces the expression of JAK/Stat targets (Socs36E) and prevents the loss of Vha100-4 expression in old guts (Fig. 3E). A similar delay in the age-related decline of the CCR was observed in flies carrying the loss of function allele hop3, while flies carrying the gain-of-function allele hopTumL (a dominant gain of function mutation of the endogenous hop gene (Hanratty and Dearolf, 1993)) exhibited accelerated CCR decline (Fig. S3I).
The specific contribution of locally and systemically derived Upds to gastric decline is likely to be complex and redundant: knockdown of Upd2 using S1106-GeneSwitch (a driver that allows strong RU486-mediated transgene induction in fatbody, but also some induction in the gut, (Alic et al., 2014; Hwangbo et al., 2004)), extended lifespan of the animal overall, but was not sufficient to prevent age-related metaplasia (Fig. S3E, F). Other Upd ligands, or other sources of Upd2, are thus likely responsible for age-related gastric metaplasia, while the lifespan extension observed upon Upd2 knockdown in fatbody is independent of gastric decline, and may be a consequence of Upd2-mediated systemic metabolic effects (Rajan and Perrimon, 2012). We observed no lifespan extension when Upd3 was knocked down using S1106 (Fig. S3E).
Mechanism of JAK/Stat–induced metaplasia
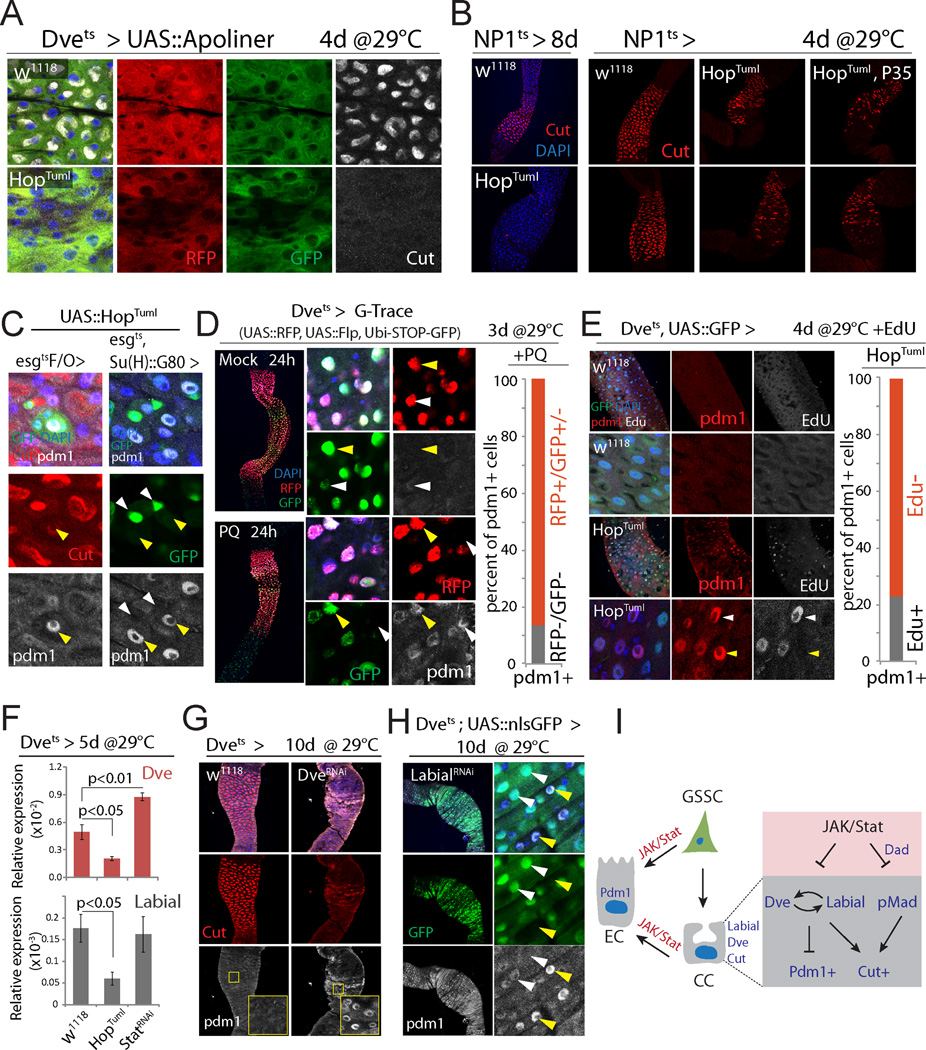
The accumulation of Pdm1+ cells in the CCR may be the result of mis-differentiation of newly formed, GSSC-derived cells, or of trans-differentiation of established CCs into Pdm1+ cells. When HopTumL was expressed in GSSCs and their daughter cells using esg::Gal4 combined with tub::Gal80ts (esgts) or in GSSC clones using the esgtsF/O lineage tracing system (Jiang et al., 2009), newly formed cells became Pdm1+ (in both the PM and the CCR), while surrounding MM cells retained Cut staining, suggesting that HopTumL can act cell autonomously in newly formed gastric cells to generate Pdm1+ polyploid cells (Fig. 4C, S4A). Pdm1+ cells were also formed when HopTumL was expressed selectively in GSSCs and not their daughter cells, using esgts combined with Su(H)::Gal80 (which inhibits Gal4 activity specifically in EBs and GBs, (Wang et al., 2014); Fig. 4C). Elevated JAK/Stat activity within GSSCs is thus sufficient to convert these cells into PM-like ISCs that produce Pdm1+ EC-like cells rather than Cut+ CCs.
Figure 4. Mechanisms of metaplasia: GSSC mis-differentiation and CC trans-differentiation.
(A) Expression of Apoliner in the CCR. No apoptosis is observed in CCs expressing HopTuml. Representative images of 7 flies per genotype.
(B) Loss of CCs (red) upon over-expression of HopTuml using NP1::Gal4ts. Co-expression of P35 does not prevent CC loss. Representative images of 9 flies per genotype.
(C) Pdm1+ cells (arrow) are generated in GSSC-derived clones (left; esgtsF/O> HopTuml, green), or when HopTuml is overexpressed only in the GSSCs (right; esg::Gal4ts with Su(H)::Gal80). Note that GFP+GSSCs (white arrow) do not express Pdm1.
(D) G-Trace expression was initiated by incubating flies at 29°C for 3d before inducing Pdm1+ cells by Paraquat (PQ) treatment. All Dve+ cells express RFP (red), while GFP expression (green) remains mosaic, indicating that Flp-mediated recombination occurs only in a subset of these cells (yellow arrowhead strong GFP, recombined; white arrowhead background GFP, non-recombined). Graph shows the ratio of Pdm1+ cells that express RFP and/or GFP. N=239 cells from 3 guts.
(E) EdU incorporation analysis to determine whether formation of Pdm1+ cells in the CCR involves DNA synthesis. Among induced Pdm1+ cells (red), 23% are EdU positive (white arrowhead), while 77% are EdU negative (yellow arrowhead). N= 126 cells from 4 guts. In w1118 CCRs, no Pdm1 or EdU is detected.
(F) qRT-PCR showing Dve and Labial expression in CCRs of indicated genotypes. Averages and s.e.m. (t-test). N=3 biological replicates.
(G) Dve knockdown results in loss of CCs (red) and generation of Pdm1+ cells (white).
(H) Labial knockdown results in loss of CCs (red) and generation of Pdm1+ cells (white). Note that in the high Pdm1+ cells, GFP (Dve::Gal4>) signal is reduced (yellow arrow) compared to low- or non-Pdm1 cells (white arrow).
(I) Model for the role of JAK/Stat signaling in promoting mis- and trans-differentiation of CCR cells.
See also Fig. S4 and S5.
However, we also observed direct trans-differentiation of established CCs into Pdm1+ cells. Since Dve::Gal4 is only expressed in differentiated, polyploid cells of the MM, it serves as an indicator of MM cell identity. Spontaneous Pdm1+ cells in the CCR of aging wild-type flies generally express low levels of Dve-driven GFP (Fig. S4B), and co-expression of HopTumL and GFP under the control of Dvets results in progressive loss of the GFP signal, and a corresponding increase in Pdm1 immunoreactivity (Fig. S4C). The formation of Pdm1+ cells is thus accompanied by suppression of Dve.
The accumulation of Pdm1+ cells upon expression of HopTumL is not accompanied by increased apoptosis (determined using Apoliner (Bardet et al., 2008), Fig. 4A, S4D), nor does co-expression of the Caspase inhibitor P35 prevent the loss of CCs (Fig. 4B, HopTumL was expressed using NP1ts in this experiment). Consistently, while apoptosis increases in the PM of aging flies, we did not observe age-related increase of apoptosis in the CCR (determined by TUNEL staining, Fig. S4E). Loss of Cut+ cells in response to JAK/Stat activation in CCs thus appears not to be driven by cell replacement, but rather by direct conversion of Cut+ cells into Pdm1+ cells. To test this hypothesis, we performed lineage tracing using G-Trace (Evans et al., 2009) and EdU incorporation experiments. In G-Trace, an inducible Gal4 (in our case Dvets) activates expression of RFP in combination with UAS::Flp. Flp will then cause excision of a STOP-FRT cassette, allowing heritable expression of a ubiquitin-promoter driven GFP. We shifted flies to 29°C to activate Dve::Gal4 for 3 days, and then exposed them to Paraquat for 24 hours to induce JAK/Stat activity (Fig. S5A). If Pdm1+ cells would be formed under these conditions directly from GSSCs (without going through a Dve+ state that is characteristic of MM cells), no Pdm1+ cell in the CCR should express GFP or RFP. However, only 10% of all Pdm1+ cells lacked GFP and RFP expression. Under stress conditions, the formation of Pdm1+ cells in the MM thus mostly involves transitioning through a Dve+ MM-like differentiated state (Fig. 4D).
We further tested whether differentiated cells in the CCR can directly convert into Pdm1+ cells using EdU incorporation. A large majority of newly formed Pdm1+ cells did not incorporate EdU even when supplied for the full period of HopTumL induction, indicating no new DNA synthesis associated with polyploidization has occurred. Cut+ CCs can thus trans-differentiate into Pdm1+ cells rather than being replaced by new cells formed from GSSCs (Fig. 4E).
Both mis-differentiation of GSSC-derived newly formed cells and trans-differentiation of established CCs can thus contribute to JAK/Stat-induced CCR metaplasia, suggesting that JAK/Stat signaling impairs molecular mechanisms that ensure formation and maintenance of CC identity. Dpp signaling is required for differentiation of CCs (Guo et al., 2013; Li et al., 2013), and the activity of Dpp signaling (pMad staining) is inhibited by JAK/Stat signaling in CCs (Fig. S5B). This inhibition was rescued by knocking down the inhibitory SMAD Daughters Against Dpp (Dad), suggesting a role for Dad in inhibiting Dpp signaling downstream of JAK/Stat activation in these cells (Fig. S5B). Supporting this notion, activation of JAK/Stat signaling led to high expression of Dad::GFP in the CCR (Fig. S5H). Inhibition of Dpp signaling, while preventing the formation of new Cut+ cells (Guo et al., 2013; Li et al., 2013), did not, however, cause the formation of Pdm1+ cells in the CCR: MARCM clones generated by ISCs homozygous for the mad12 loss of function allele, while containing Pdm1+ cells in the PM, did not contain such cells in the CCR (Fig. S5C). Thus, inhibition of Dpp signaling, which was also seen in old CCRs (Fig. S5G), contributes to the loss of CCs, but not to the formation of Pdm1+ cells. Furthermore, knocking down Dad was not sufficient to rescue the UAS::HopTumL-induced loss of CCs (Fig. S5B), indicating that other mechanisms must act in parallel to perturb CC maintenance in JAK/Stat gain-of-function conditions.
The transcription factors Dve and Labial are both expressed specifically in polyploid cells, including CCs, of the MM. Consistent with the suppression of Dve expression in Pdm1+ CCs (Fig. S4C), Dve transcripts in the CCR are reduced or increased when HopTuml or StatRNAi are expressed, respectively (Fig. 4F). Similarly, Labial expression is repressed in GSSC lineages expressing HopTuml (esgtsF/O; Fig. 4F, S5D). Knockdown of either Dve or Labial using Dvets results in the formation of Pdm1+ cells and loss of CCs (Fig. 4G, H), and Labial-deficient Pdm1+ cells exhibit low Dve::Gal4 activity, while knockdown of Dve reduces Labial expression (Fig. 4H, S5E). Knockdown of Dve did not, however, affect Dpp signaling as determined by pMad staining (Fig. S5F).
We propose that CC identity is established and maintained by the co-dependent expression of Dve and Labial, and that excessive JAK/Stat activation promotes trans-differentiation of CCs by independently perturbing the Labial-Dve network and Dpp signaling (Fig. 4I).
Inhibiting JAK/Stat signaling promotes gut homeostasis and extends lifespan
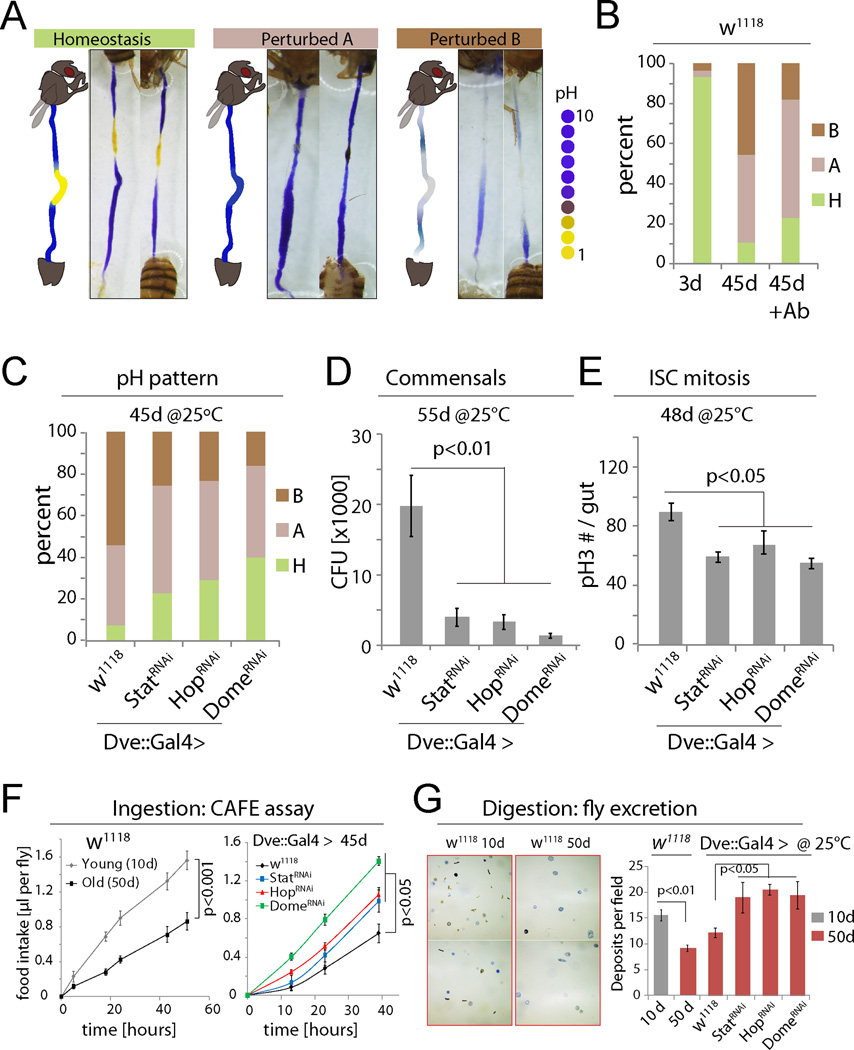
To test the importance of JAK/Stat–mediated metaplasia in the development of age-related intestinal dysfunction, reduced JAK/Stat activity in the CCR and assessed age-related phenotypes in the gut. More than 90% of wild-type flies lose pH homeostasis with age (Fig. 5A, B, S6A). However, the observed phenotypes are not uniform: 44% of 45d old flies had a highly basic gut with no acidic region, while 46% exhibited weak staining throughout the gut (Fig. 5A, B, S6A). While the loss of the acidic region was expected based on the observed decline of the CCR, the loss of staining in other flies was surprising, and further analysis suggested that it is caused by the age-related expansion of acid-producing commensals, like Lactobacillus: when old flies were treated with antibiotics, the proportion of flies with reduced staining declined, while flies with a more basic gut lumen increased (Fig. 5B, S6A). Furthermore, feeding young flies Lactobacilli cultured from fly guts (these cultures themselves have a pH of around 4) resulted in an intestinal pH pattern that was similar to old flies with increased intestinal acidity (Fig. S6B; Lactobacillus readily colonized these guts, Fig. S6C). Ingestion of a blue food dye confirmed that these flies took in similar amounts of food as sucrose-fed animals (Fig. S6B). The loss of staining in old guts is thus due to Lactobacillus-induced luminal changes or a bacteria-induced barrier dysfunction.
Figure 5. Inhibiting JAK/Stat signaling in the CCR promotes gut function and homeostasis.
(A) GI tracts of flies fed the pH indicator Bromophenol Blue. Three phenotypic categories can be distinguished: homeostasis, a well-defined acidic CCR is flanked by basic AM and PM; ‘Perturbed A’, the acidic region is lost and the whole gut is basic; ‘Perturbed B’, the strongly acidic region is lost, while the rest of the gut also becomes less basic (weak blue).
(B) Quantification of the three phenotypes in flies of indicated conditions. +Ab: Antibiotic treatment. N=28, 57, 39. Representative of 3 independent experiments.
(C–E) Rescue of pH homeostasis (C; N=13, 25, 17, 39), commensal dysbiosis (D; N=10 each), and epithelial dysplasia (E; pH3+ cells in the gut, N= 12, 20, 12, 12, 12, 20, 12, 12) in old flies expressing StatRNAi, HopRNAi or DomeRNAi in the CCR. CFU: colony forming units. Representative of 3 independent experiments.
(F) Food intake measured using CAFÉ assay. Averages and s.e.m. (t-test). N=18 flies from 6 replicates (w1118) or 21 flies from 7 replicates (other genotypes).
(G) Excretion from flies fed Bromophenol blue. Images of deposits and quantification of deposit numbers are shown. Excretions are quantified in 8 to 24 fields for each group of 12 flies. Averages and s.e.m. (t-test). N= 24, 24, 8, 12, 12, 12 fields.
See also Fig. S6
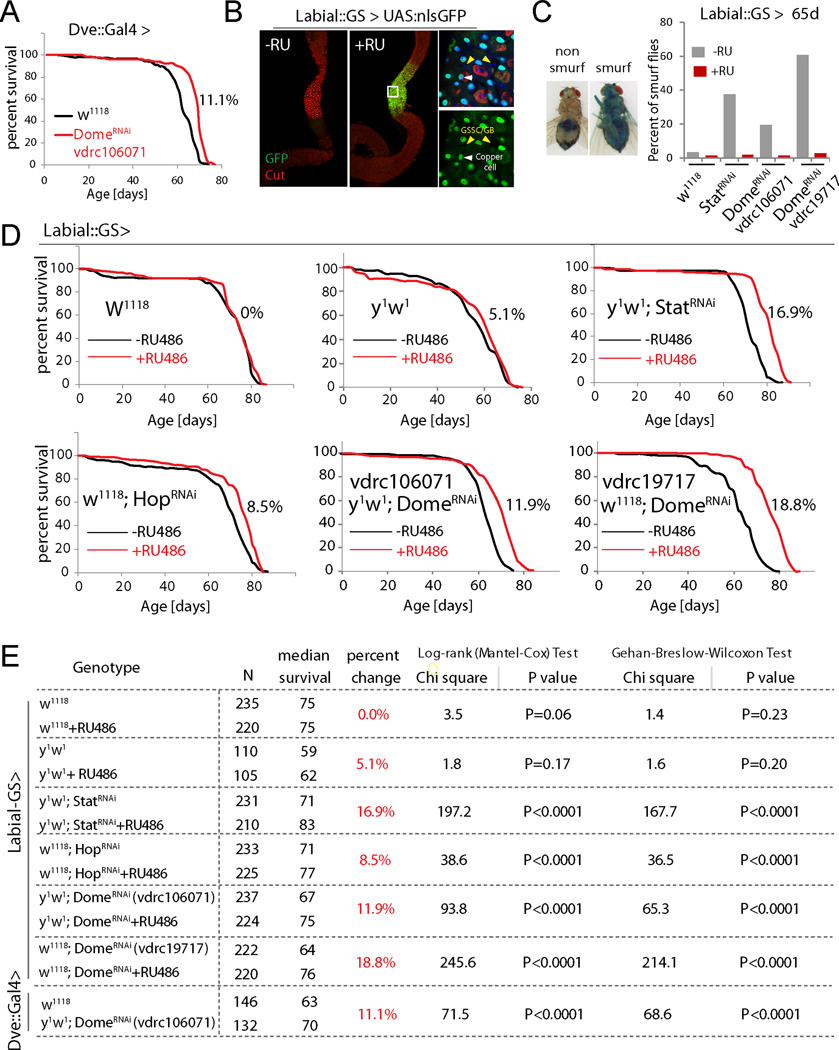
Knocking down JAK/Stat signaling components using Dve::Gal4 restored pH homeostasis, limited commensal dysbiosis, reduced dysplasia (Fig. 5C–E), and prevented the age-related decline in both food intake (CAFÉ assay, (Ja et al., 2007)) and excretion (Fig. 5F, G, S6D, E). Flies in which Dome was knocked down using Dve::Gal4 further lived significantly longer than controls (Fig. 6A). We confirmed the lifespan extension by limiting JAK/Stat signaling in the CCR using a newly generated RU486-responsive Geneswitch (McGuire et al., 2004b) driver based on labial regulatory regions (Labial::GS; Fig. S7A) (Jenett et al., 2012). In response to RU486, this driver selectively induces UAS-linked transgene expression in the MM and is not expressed in other regions of the gut or in other tissues (Fig. 6B, S7B). This driver thus allows selective repression of JAK/STAT components in the MM in a temporally controlled manner and allows for comparison of genetically identical sibling populations exposed to RU486 or vehicle. When Stat, Hop or Dome were knocked down throughout adulthood using this driver, the percentage of old animals exhibiting a breakdown of intestinal barrier function (‘Smurf’ assay (Rera et al., 2012)) was reduced, and lifespan was consistently and significantly increased after RU486 supplementation (Fig. 6C–E, S7C). Reducing JAK/Stat activity in the CCR also extended lifespan of flies cultured on antibiotic food (Fig. S7D), suggesting that maintenance of GI compartmentalization impacts lifespan not only by improving microbiota homeostasis, but has additional beneficial effects. Flies cultured on antibiotic food generally have a longer lifespan than flies cultured on conventional food (Fig. S7C and S7D), consistent with other studies (Brummel et al., 2004; Clark et al., 2015).
Figure 6. Inhibiting JAK/Stat signaling in the CCR extends lifespan (See also Fig. S7).
(A) Demography of flies expressing DomeRNAi in the CCR (w1118 as control). The percent change of median lifespan is indicated. All flies are female.
(B) Characterization of Labial-GeneSwitch. In response to RU486, UAS::GFP expression is observed in the CCR specifically. High magnification (right) shows that this expression is in differentiated polyploid cells (including CCs, white arrowhead) but not in diploid cells (GSSCs and GBs, yellow arrowhead). Representative images of 7 flies for each condition.
(C) Smurf assay for gut permeability of 65d old flies. Percent of Smurf flies are shown. N= 137, 139, 122, 121, 123, 147, 128, 143.
(D) Demography of flies expressing StatRNAi, HopRNAi or two different DomeRNAi constructs under the control of Labial::GS, with w1118 and y1w1 as control. Graphs are combined from independent populations (only one population for y1w1) of F1 progeny from Labial::GS crossed to the indicated transgenes or to w1118 and y1w1 (control). The percent changes of median lifespan are indicated. All flies are female. Survival data was analyzed using GraphPad Prism.
(E) Summary of parameters and statistics of the demography shown in (A) and (D). Survival data was analyzed using GraphPad Prism.
See also Fig. S7
Discussion
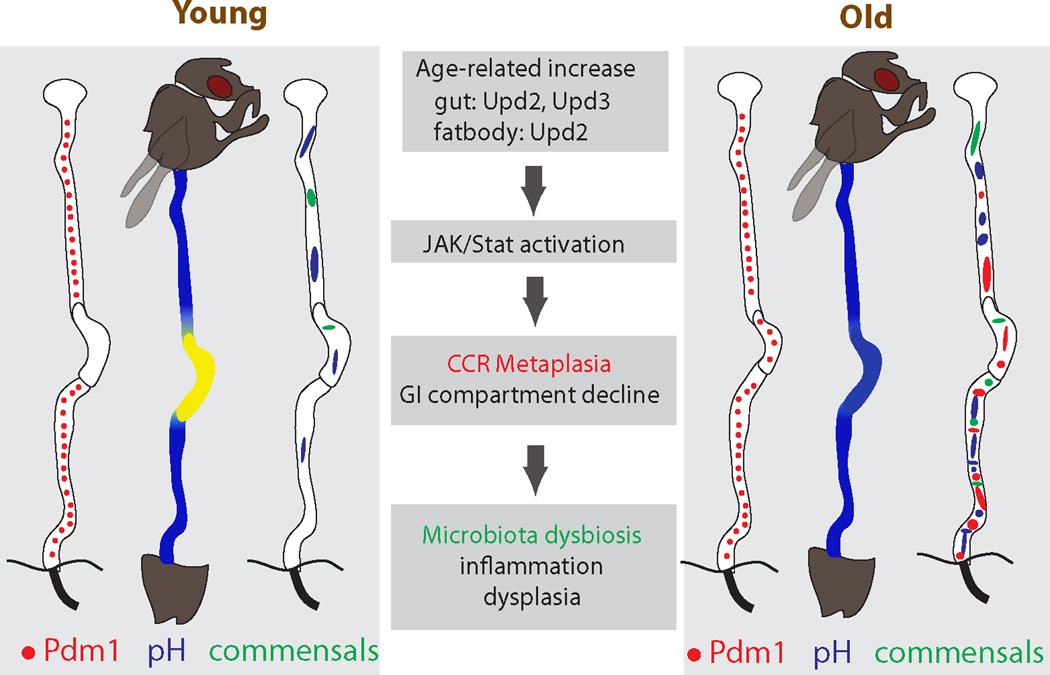
Our results identify a critical role for the gastric region in managing commensal compartmentalization, composition and load in the GI tract of adult flies. The importance of this function is reflected by the significant deleterious consequences of age-related epithelial metaplasia. Metaplasia of the gastric epithelium, caused by chronic inflammation (in the form of JAK/Stat activation), thus emerges as the origin of age-related pathophysiological changes in the gastrointestinal tract of Drosophila, highlighting the role of low-level inflammation as a driver of age-related pathologies (Fig. 7). This observation, as well as insight from previous studies, provides a model for the molecular events driving the progression of age-related pathological changes in the intestinal epithelium (Fig. 7) (Biteau et al., 2010b; Clark et al., 2015; Guo et al., 2014; Rera et al., 2012).
Figure 7. Origin and pathophysiological consequences of age-related gastric metaplasia.
Age-related increase of JAK/Stat activity, induced by increased cytokines from the gut and/or fat body, causes metaplasia in the CCR. This results in pH imbalance, promoting commensal dysbiosis, inflammation and dysplasia in the old intestine.
Previous studies have linked age-related immunosenescence to microbiota dysbiosis and epithelial dysplasia, and have identified the loss of barrier function as a significant step triggering changes in both load and composition of the intestinal microbiota, thus increasing mortality (Clark et al., 2015; Guo et al., 2014). Our results introduce gastric metaplasia as an initiating age-related dysfunction that subsequently triggers the phenotypes discussed above. We propose that JAK/Stat-induced gastric metaplasia causes pH changes, resulting in dysbiosis and innate immune deregulation, and triggering a secondary inflammatory response that damages the epithelium and causes excessive ISC activity, leading to dysplasia and shortening lifespan (Guo et al., 2014). The lifespan extension observed when JAK/STAT was reduced in the CCR of sterile flies indicates broader beneficial consequences of this perturbation that may result from improved food digestion and nutrient assimilation. Furthermore, since microbes also have a beneficial effect on fly nutrition (Yamada et al., 2015), it is likely that the metabolic consequences of age-related gastric decline are complex and provide interesting grounds for future studies.
In humans, Barrett’s metaplasia, in which the esophageal squamous epithelium acquires properties that are reminiscent of the gastric or intestinal epithelium, is believed to be a cause of esophageal adenocarcinomas (Dvorak et al., 2011; Peters and Avisar, 2010), and metaplasia of the gastric mucosa has been associated with the progression from inflammation to dysplasia and carcinogenesis (Correa and Houghton, 2007). Barrett’s metaplasia and malignant transformation of intestinal progenitor cells has been linked to inflammation and JAK/Stat activation (Liu et al., 2013; Nguyen et al., 2010; Ullman and Itzkowitz, 2011), and in vertebrate airway epithelia, JAK/Stat activating inflammatory cytokines have been implicated in asthma-induced metaplasias (Ren et al., 2013). Pre-malignant intestinal-type metaplasias in the gastric region are common in the general population and their incidence increases with age (Zullo et al., 2012), and our results suggest that such metaplasias occur as a consequence of basal inflammation in the aging organism. Accordingly, age-related loss of tissue homeostasis and cancer progression is associated with an inflammatory state (Grivennikov et al., 2010; Mantovani, 2009; Shaw et al., 2013). Based on this model, it can be anticipated that interventions targeting basal inflammation, and thus preventing chronic activation of JAK/Stat signaling, may significantly promote tissue maintenance, regenerative capacity, and tissue homeostasis, ultimately extending lifespan.
Experimental procedures
Fly lines and husbandry
Immunostaining, TUNEL staining, and Microscopy
Female guts were dissected in 1XPBS, fixed for 45 minutes at 25°C in fixative, washed for 1 hour at 4°C in washing buffer, and then incubated at 4°C in primary antibodies overnight and secondary antibodies for 4 hours. TUNEL staining was performed using In Situ Cell Death Detection Kit (Roche, 12156792910). Images were taken on a Zeiss LSM 700 confocal microscope and processed using Adobe Photoshop, Illustrator and Image J.
See Supplemental Experimental Procedures for details about fixative, washing buffer and antibodies.
pH indicator, blue dye, paraquat treatment, and Lactobacillus feeding
100ul of 2% Bromophenol blue sodium (pH indicator, Sigma), or 5% sucrose ± 80 mM paraquat (methyl viologen, Sigma) was added to a food vial and pipet tip was used to poke 4–6 holes in the food to allow full absorption. Flies were fed for overnight. For Lactobacillus feeding, flies were fed 400 ul of concentrated bacteria (OD100, in 5% sucrose) mixed with either 100ul 2% pH indicator or 2.5% blue dye (FD&C Blue 1, SENSIENT) in filter paper for overnight. During dissection, the fly head and posterior cuticle was left intact to prevent dye leakage. Images were taken immediately after each gut was dissected. Long-term exposure of flies with pH indicator on CO2 pad will disturb normal pH patterns. Edu incorporation
The Click-iT EdU Imaging Kit from Invitrogen was applied. Briefly, 100ul EdU solution (100uM, diluted with water from 10mM stock) was added to a food vial (pipet tip was used to poke 4–6 holes in the food to allow full absorption of the solution). Flies were fed EdU food at 29°C for 4 days.
Demography and axenic fly culture
The procedure was performed as described (Guo et al., 2014). See Supplemental Experimental Procedures for details.
esgtsF/O and MARCM Clone induction
Because of the intrinsic quiescence of gastric stem cells, the frequency of clone formation in the CCR is low. Double heat-shock (45min at 37°C, 3h at 25°C, then 45min at 37°C) was applied to induce enough clones for analysis.
See Supplemental Experimental Procedures for details.
Commensal quantification and selective plates
The procedure was performed as described (Guo et al., 2014). In brief, dissected guts were homogenized in 200 ml sterile 1XPBS and plated onto selective plates, which were incubated at 29°C for 48–72 hours before the colony forming units were quantified. See Supplemental Experimental Procedures for recipes of selective plates.
Generation of Labial-GeneSwitch
A collection of Janelia-Gal4 lines with varied labial promoter regions were screened (Jenett et al., 2012) for their expression in the intestine, and one line (BL49323) was found to be expressed most specifically in the CCR. The 3388 bp promoter region corresponding to that line was amplified (primers: 5’-CCACCCCCTGGAATCGGTAATTTCT and 5’-GAGCGCGTGTGACAAGATGGATAAA), and inserted into pP[UAS-GeneSwitch] using MluI and NotI sites (Osterwalder et al., 2001). Transgenic animals were generated in the w1118 background using standard P-element transgenesis (GENETIC SERVICES, Inc.).
16S rDNA sequencing and PCA analysis
See Supplemental Experimental Procedures for the preparation of sequencing library.
Illumina Miseq paired-end (2×300bp) sequencing was performed and Miseq Reporter Software was used for primary analysis and classification based on Greengenes database (http://greengenes.lbl.gov/). We noticed that for samples with low numbers of bacteria, e.g. young wild-type guts, there were high percentages of unclassified species. Further analysis (by Tal Ronnen Oron) of DNA sequences responsible for these unclassified species revealed that these DNA sequences were from the Drosophila genome, presumably due to non-specific amplification during the preparation of 16S sequencing libraries; these sequences were excluded in subsequent analysis). The percentage of each classified bacterium at the species level was used for principle component analysis (PCA) using Matlab (princomp command), and Prism software was used for plotting the data.
Commensal DNA concentration
RFP-tagged Lactobacillus
The plasmid pLEM415-ldhL-mRFP1 (Bao et al., 2013) was used for transformation of one strain of commensal Lactobacillus, which was isolated and cultured from wild type flies. The procedure of competent cell preparation and electrical transformation has been described (Bao et al., 2013). The transformed strain was later identified as Lactobacillus plantarum by sequencing the 16S rRNA gene.
RT-PCR and primer sequences
Cafe assay and fly excretion measurement
The Cafe and fly excretion assays were performed as described (Deshpande et al., 2014) and (Cognigni et al., 2011), respectively. See Supplemental Experimental Procedures for details.
Statistical methods
Statistical Analysis was performed using GraphPad Prism5 and Microsoft Excel. Statistical methods used and sample sizes are listed in Figure Legends. Sample sizes were chosen empirically based on observed effect sizes.
Supplementary Material
Acknowledgments
This work was funded by the National Institute on Aging (R01 AG028127), the National Institute on General Medical Sciences (R01 GM100196), and the American Federation for Aging Research (Breakthroughs in Gerontology award to H.J.). We would like to thank the Vienna Drosophila RNAi Center and the Bloomington Stock Center for flies, and Developmental Studies Hybridoma Bank for antibodies. We thank Rachel Brem and Tal Ronnen Oron for bioinformatics support, Jason Karpac for valuable comments on the manuscript, and Jasper lab members for discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
H.L. and H.J. designed and conceived the study, H.L. and Y. Q. performed all experiments, H.J. and H.L. analyzed data and wrote the manuscript.
References
- Alic N, Giannakou ME, Papatheodorou I, Hoddinott MP, Andrews TD, Bolukbasi E, Partridge L. Interplay of dFOXO and two ETS-family transcription factors determines lifespan in Drosophila melanogaster. PLoS genetics. 2014;10:e1004619. doi: 10.1371/journal.pgen.1004619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyaz A, Jasper H. Intestinal inflammation and stem cell homeostasis in aging. Frontiers in cellular and infection microbiology. 2013;3:98. doi: 10.3389/fcimb.2013.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach EA, Ekas LA, Ayala-Camargo A, Flaherty MS, Lee H, Perrimon N, Baeg GH. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene expression patterns : GEP. 2007;7:323–331. doi: 10.1016/j.modgep.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Bao S, Zhu L, Zhuang Q, Wang L, Xu PX, Itoh K, Holzman IR, Lin J. Distribution dynamics of recombinant Lactobacillus in the gastrointestinal tract of neonatal rats. PloS one. 2013;8:e60007. doi: 10.1371/journal.pone.0060007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardet PL, Kolahgar G, Mynett A, Miguel-Aliaga I, Briscoe J, Meier P, Vincent JP. A fluorescent reporter of caspase activity for live imaging. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13901–13905. doi: 10.1073/pnas.0806983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, Bartfeld S, Clevers H. Tissue-resident adult stem cell populations of rapidly self-renewing organs. Cell Stem Cell. 2010;7:656–670. doi: 10.1016/j.stem.2010.11.016. [DOI] [PubMed] [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell. 2008;3:442–455. doi: 10.1016/j.stem.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H. Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell Stem Cell. 2011;9:402–411. doi: 10.1016/j.stem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Karpac J, Hwangbo D, Jasper H. Regulation of Drosophila lifespan by JNK signaling. Experimental gerontology. 2010a;46:349–354. doi: 10.1016/j.exger.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Karpac J, Supoyo S, DeGennaro M, Lehmann R, Jasper H. Lifespan extension by preserving proliferative homeostasis in Drosophila. PLoS Genet. 2010b;6:e1001159. doi: 10.1371/journal.pgen.1001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum JE, Fischer CN, Miles J, Handelsman J. Frequent replenishment sustains the beneficial microbiome of Drosophila melanogaster. MBio. 2013;4:e00860–e00813. doi: 10.1128/mBio.00860-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick NA, Buchon N, Lemaitre B. Microbiota-induced changes in drosophila melanogaster host gene expression and gut morphology. MBio. 2014;5:e01117–e01114. doi: 10.1128/mBio.01117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummel T, Ching A, Seroude L, Simon AF, Benzer S. Drosophila lifespan enhancement by exogenous bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12974–12979. doi: 10.1073/pnas.0405207101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes & development. 2009;23:2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Osman D, David FP, Fang HY, Boquete JP, Deplancke B, Lemaitre B. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell reports. 2013;3:1725–1738. doi: 10.1016/j.celrep.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Claesson MJ, Cusack S, O'Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, Marchesi JR, Falush D, Dinan T, Fitzgerald G, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, Cusack S, Harris HM, Coakley M, Lakshminarayanan B, O'Sullivan O, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- Clark RI, Salazar A, Yamada R, Fitz-Gibbon S, Morselli M, Alcaraz J, Rana A, Rera M, Pellegrini M, Ja WW, et al. Distinct Shifts in Microbiota Composition during Drosophila Aging Impair Intestinal Function and Drive Mortality. Cell reports. 2015;12:1656–1667. doi: 10.1016/j.celrep.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classen AK, Bunker BD, Harvey KF, Vaccari T, Bilder D. A tumor suppressor activity of Drosophila Polycomb genes mediated by JAK-STAT signaling. Nature genetics. 2009;41:1150–1155. doi: 10.1038/ng.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cognigni P, Bailey AP, Miguel-Aliaga I. Enteric neurons and systemic signals couple nutritional and reproductive status with intestinal homeostasis. Cell metabolism. 2011;13:92–104. doi: 10.1016/j.cmet.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa P, Houghton J. Carcinogenesis of Helicobacter pylori. Gastroenterology. 2007;133:659–672. doi: 10.1053/j.gastro.2007.06.026. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande SA, Carvalho GB, Amador A, Phillips AM, Hoxha S, Lizotte KJ, Ja WW. Quantifying Drosophila food intake: comparative analysis of current methodology. Nature methods. 2014;11:535–540. doi: 10.1038/nmeth.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil RR. Copper cells and stomach acid secretion in the Drosophila midgut. The international journal of biochemistry & cell biology. 2004;36:745–752. doi: 10.1016/j.biocel.2003.07.004. [DOI] [PubMed] [Google Scholar]
- Dvorak K, Goldman A, Kong J, Lynch JP, Hutchinson L, Houghton JM, Chen H, Chen X, Krishnadath KK, Westra WM. Molecular mechanisms of Barrett's esophagus and adenocarcinoma. Annals of the New York Academy of Sciences. 2011;1232:381–391. doi: 10.1111/j.1749-6632.2011.06062.x. [DOI] [PubMed] [Google Scholar]
- Evans CJ, Olson JM, Ngo KT, Kim E, Lee NE, Kuoy E, Patananan AN, Sitz D, Tran P, Do MT, et al. G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila. Nature methods. 2009;6:603–605. doi: 10.1038/nmeth.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Karpac J, Tran SL, Jasper H. PGRP-SC2 Promotes Gut Immune Homeostasis to Limit Commensal Dysbiosis and Extend Lifespan. Cell. 2014;156:109–122. doi: 10.1016/j.cell.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Driver I, Ohlstein B. Injury-induced BMP signaling negatively regulates Drosophila midgut homeostasis. J Cell Biol. 2013;201:945–961. doi: 10.1083/jcb.201302049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanratty WP, Dearolf CR. The Drosophila Tumorous-lethal hematopoietic oncogene is a dominant mutation in the hopscotch locus. Molecular & general genetics : MGG. 1993;238:33–37. doi: 10.1007/BF00279527. [DOI] [PubMed] [Google Scholar]
- Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- Ja WW, Carvalho GB, Mak EM, de la Rosa NN, Fang AY, Liong JC, Brummel T, Benzer S. Prandiology of Drosophila and the CAFE assay. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8253–8256. doi: 10.1073/pnas.0702726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell reports. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzelius J, Naumann SK, Loza-Coll MA, Chan JS, Dutta D, Oberheim J, Glasser C, Southall TD, Brand AH, Jones DL, et al. Escargot maintains stemness and suppresses differentiation in Drosophila intestinal stem cells. EMBO J. 2014;33:2967–2982. doi: 10.15252/embj.201489072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WC, Beebe K, Sudmeier L, Micchelli CA. Adenomatous polyposis coli regulates Drosophila intestinal stem cell proliferation. Development. 2009;136:2255–2264. doi: 10.1242/dev.035196. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Miguel-Aliaga I. The digestive tract of Drosophila melanogaster. Annual review of genetics. 2013;47:377–404. doi: 10.1146/annurev-genet-111212-133343. [DOI] [PubMed] [Google Scholar]
- Li H, Qi Y, Jasper H. Dpp signaling determines regional stem cell identity in the regenerating adult Drosophila gastrointestinal tract. Cell Rep. 2013;4:10–18. doi: 10.1016/j.celrep.2013.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WS, Huang CW, Song YS, Yen JH, Kuo PC, Yeh SR, Lin HY, Fu TF, Wu MS, Wang HD, et al. Reduced Gut Acidity Induces an Obese-Like Phenotype in Drosophila melanogaster and in Mice. PloS one. 2015;10:e0139722. doi: 10.1371/journal.pone.0139722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Jiang M, Lu Y, Chen H, Sun J, Wu S, Ku WY, Nakagawa H, Kita Y, Natsugoe S, et al. Sox2 cooperates with inflammation-mediated Stat3 activation in the malignant transformation of foregut basal progenitor cells. Cell stem cell. 2013;12:304–315. doi: 10.1016/j.stem.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A. Cancer: Inflaming metastasis. Nature. 2009;457:36–37. doi: 10.1038/457036b. [DOI] [PubMed] [Google Scholar]
- Marianes A, Spradling AC. Physiological and stem cell compartmentalization within the Drosophila midgut. eLife. 2013;2:e00886. doi: 10.7554/eLife.00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Mao Z, Davis RL. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Science's STKE : signal transduction knowledge environment. 2004a;2004:pl6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Roman G, Davis RL. Gene expression systems in Drosophila: a synthesis of time and space. Trends in genetics : TIG. 2004b;20:384–391. doi: 10.1016/j.tig.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Nguyen GH, Schetter AJ, Chou DB, Bowman ED, Zhao R, Hawkes JE, Mathe EA, Kumamoto K, Zhao Y, Budhu A, et al. Inflammatory and microRNA gene expression as prognostic classifier of Barrett's-associated esophageal adenocarcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:5824–5834. doi: 10.1158/1078-0432.CCR-10-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman D, Buchon N, Chakrabarti S, Huang YT, Su WC, Poidevin M, Tsai YC, Lemaitre B. Autocrine and paracrine unpaired signaling regulate intestinal stem cell maintenance and division. Journal of cell science. 2012;125:5944–5949. doi: 10.1242/jcs.113100. [DOI] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JH, Avisar N. The molecular pathogenesis of Barrett's esophagus: common signaling pathways in embryogenesis metaplasia and neoplasia. Journal of gastrointestinal surgery : official journal of the Society for Surgery of the Alimentary Tract. 2010;14(Suppl 1):S81–S87. doi: 10.1007/s11605-009-1011-7. [DOI] [PubMed] [Google Scholar]
- Rajan A, Perrimon N. Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell. 2012;151:123–137. doi: 10.1016/j.cell.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Shah TA, Ustiyan V, Zhang Y, Shinn J, Chen G, Whitsett JA, Kalin TV, Kalinichenko VV. FOXM1 promotes allergen-induced goblet cell metaplasia and pulmonary inflammation. Molecular and cellular biology. 2013;33:371–386. doi: 10.1128/MCB.00934-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M, Clark RI, Walker DW. Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:21528–21533. doi: 10.1073/pnas.1215849110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, Bae JW, Lee DG, Shin SC, Ha EM, Lee WJ. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- Shaw AC, Goldstein DR, Montgomery RR. Age-dependent dysregulation of innate immunity. Nature reviews Immunology. 2013;13:875–887. doi: 10.1038/nri3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand M, Micchelli CA. Quiescent gastric stem cells maintain the adult Drosophila stomach. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17696–17701. doi: 10.1073/pnas.1109794108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman TA, Itzkowitz SH. Intestinal inflammation and cancer. Gastroenterology. 2011;140:1807–1816. doi: 10.1053/j.gastro.2011.01.057. [DOI] [PubMed] [Google Scholar]
- Wang L, Zeng X, Ryoo HD, Jasper H. Integration of UPRER and oxidative stress signaling in the control of intestinal stem cell proliferation. PLoS genetics. 2014;10:e1004568. doi: 10.1371/journal.pgen.1004568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada R, Deshpande SA, Bruce KD, Mak EM, Ja WW. Microbes Promote Amino Acid Harvest to Rescue Undernutrition in Drosophila. Cell reports. 2015 doi: 10.1016/j.celrep.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Rasmussen A, Lee S, Agaisse H. The UPD3 cytokine couples environmental challenge and intestinal stem cell division through modulation of JAK/STAT signaling in the stem cell microenvironment. Developmental biology. 2013;373:383–393. doi: 10.1016/j.ydbio.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zullo A, Hassan C, Romiti A, Giusto M, Guerriero C, Lorenzetti R, Campo SM, Tomao S. Follow-up of intestinal metaplasia in the stomach: When, how and why. World journal of gastrointestinal oncology. 2012;4:30–36. doi: 10.4251/wjgo.v4.i3.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.