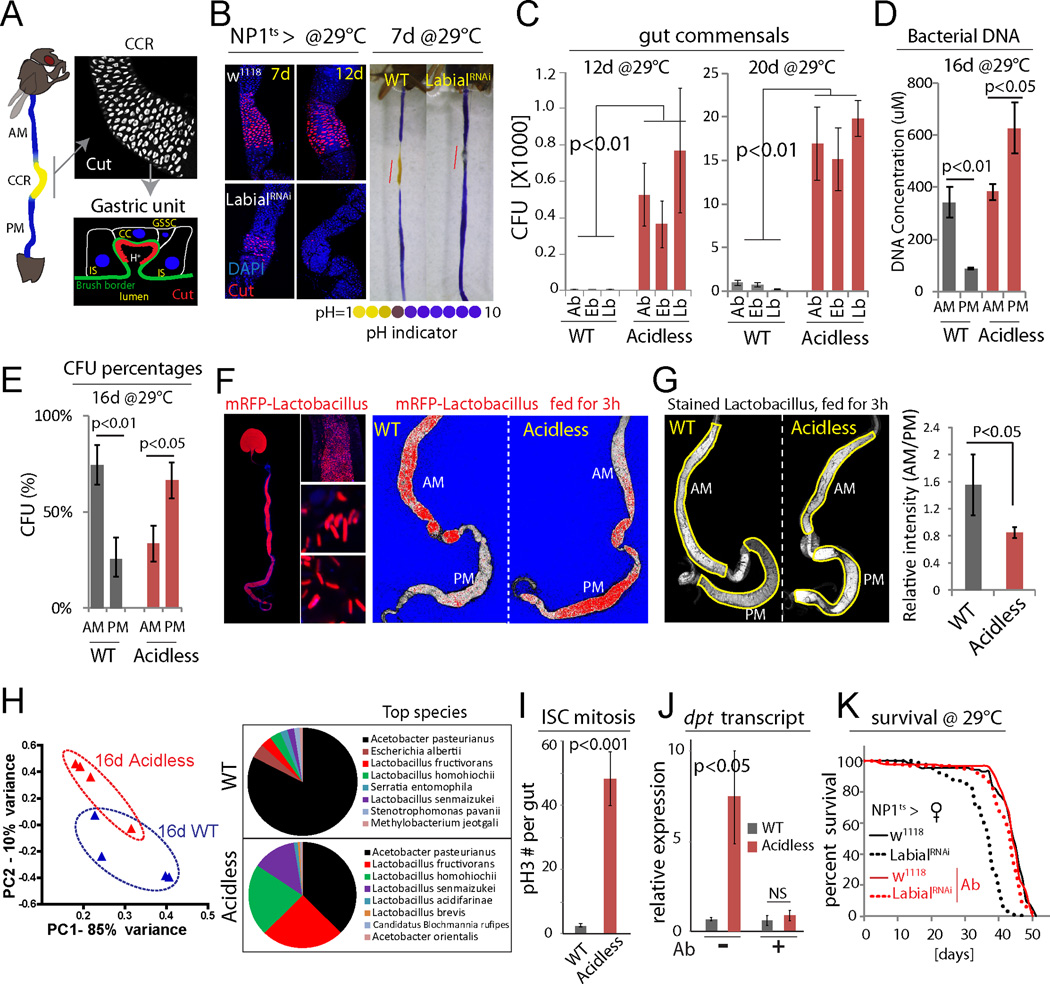
Figure 1. Intestinal compartmentalization impacts gut microbiota.
(A) Left: the Drosophila GI tract consists of anterior midgut (AM), middle midgut (MM) with copper cell region (CCR), and posterior midgut (PM). Right: anti-Cut (white) labels acid-producing copper cells (CCs). A gastric unit. IS: interstitial cell, GSSC: gastric stem cell.
(B) Expression of LabialRNAi results in loss of CCs. Acidic regions (yellow) indicated by pH indicator Bromophenol blue. Representative images of 3 independent experiments. N=7 for each genotype and time point.
(C) Commensal numbers in 12d and 20d old WT (NP1ts>w1118) and acidless flies (NP1ts>LabialRNAi). Colony-forming units (CFUs) of three common commensal phylotypes (Acetobacter, Ab; Enterobacter, Eb; Lactobacillus, Lb) are quantified using selective plates. Averages and s.e.m. (t-test), N=6, 6, 6, 10, 10, 10 (both 12d and 20d). Representative from 3 independent experiments.
(D) Bacterial DNA concentration of the AM and PM from 16d old WT and acidless flies. Averages and s.e.m. (t-test), N=3×10 guts each.
(E) Commensal distribution in 16d old WT and acidless guts. Percent averages of colony-forming units (CFUs) in AMs and PMs are shown. N=12, 11. Averages and s.e.m. (t-test).
(F) Distribution of RFP-tagged Lactobacillus plantarum in 16d old WT and acidless flies. Left: representative images of RFP-tagged Lactobacillus in the gut. Right: representative saturation images of WT and acidless guts with RFP-tagged Lactobacillus. Saturated signal red, un-saturated signal white and background blue. False color images generated from confocal images in ZEN. Representative of 3 independent experiments.
(G) Distribution of stained Lactobacillus (by SYTO 9) in 16d old WT and acidless flies. Relative fluorescent intensity is quantified. N=6, 9. Averages and s.e.m. (t-test).
(H) Commensal composition of 16d old WT and acidless guts. Principal component analysis (PCA) based on percentages of classified bacterial species determined by 16S rRNA sequencing. Pie charts show top bacterial species. For PCA, each dot represents 10 guts; for pie charts, each represents the average of four samples (4×10 guts).
(I) Quantification of pH3+ cells in guts of 16d old flies at 29°C. Averages and s.e.m. (t-test); N=12, 11. Data are representative of 3 independent experiments.
(J) qRT-PCR showing dpt expression of 16d old flies on normal or antibiotic (Ab) food. Averages and s.e.m. (t-test) of N=3 biological replicates.
(K) Lifespan of acidless flies compared to WT controls on normal or antibiotic (Ab) food. Without antibiotics: WT N=88, acidless N=93, Log-rank P value <0.0001, 17.8% change of median lifespan; Antibiotics: WT N=110, acidless N=100, Log-rank P value =0.02, no change of median lifespan.
See also Fig. S1