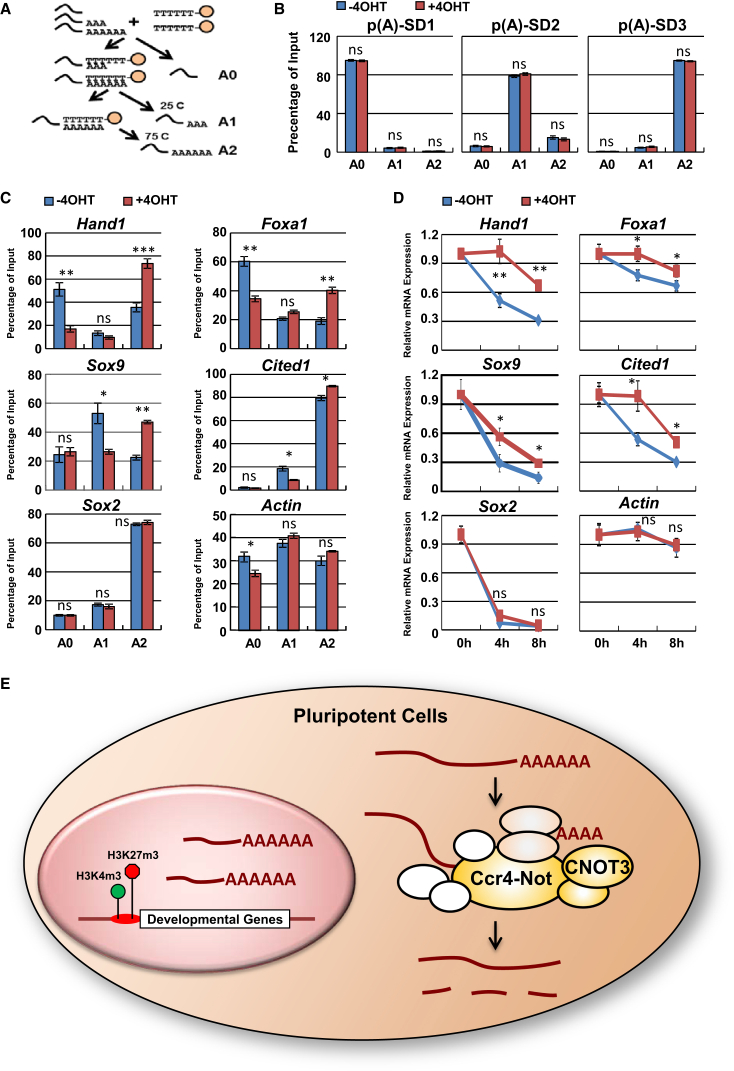
Figure 6.
Cnot3 Deletion Increases Differentiation Gene mRNA Poly(A) Tail Lengths
(A) Schematic drawing for mRNA fractionation based on poly(A) tail length.
(B) Validation of the oligo(dT) fractionation method. Cnot3 cKO ESCs were treated with (KO) or without 4-OHT (WT) for 48 hr. The poly(A) standards were mixed with total RNAs extracted from the cells, and RNAs were fractionated by oligo(dT) beads. The distribution of each standard in each fraction (A0, A1, A2) was determined by qRT-PCR and plotted as mean ± SEM from three independent experiments.
(C) Measurements of poly(A) tail length for the indicated genes by oligo(dT) fractionation.
(D) Examination of mRNA stability for the indicated genes. Cnot3 cKO ESCs were treated with (KO) or without 4-OHT (WT) for 48 hr. Actinomycin D was added to the cells, and mRNA level was measured by qRT-PCR at the indicated time points. Relative expression values were plotted as mean ± SEM from three independent experiments.
(E) Proposed model. CNOT3-dependent differentiation gene mRNA deadenylation and degradation plays a critical role in the maintenance of the pluripotent state.
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; ns, non-significant.