Figure 1.
Small Molecules Enhance NEUROG2 Chromatin Occupancy and Activate Synergistic Transcription
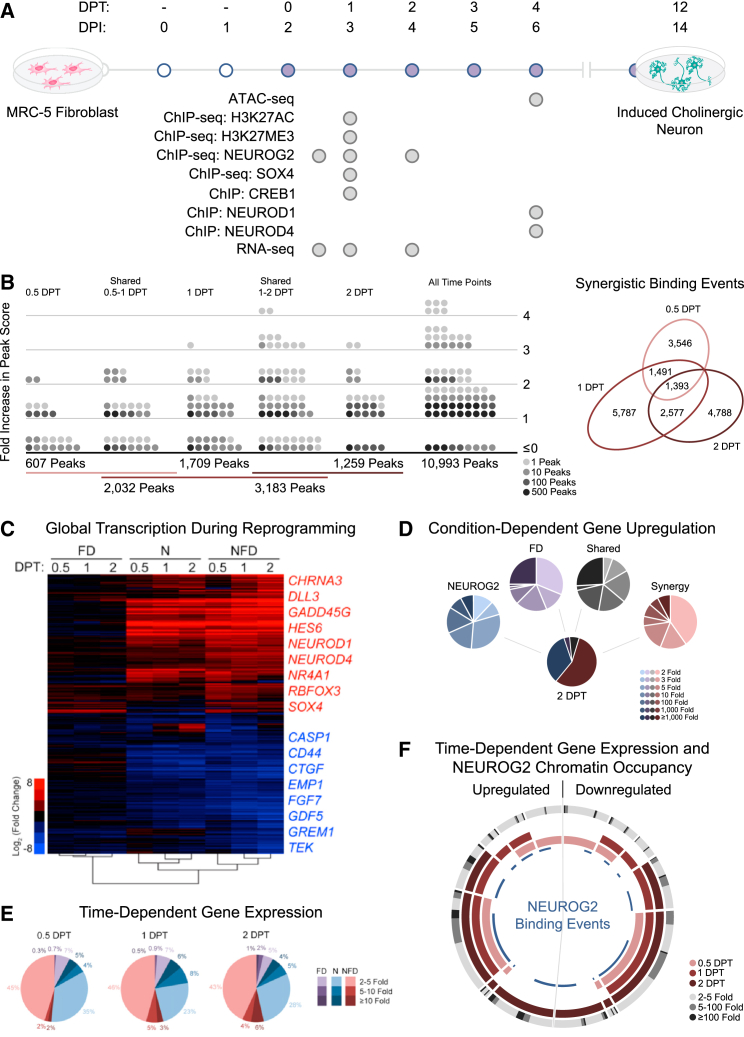
(A) Stepwise timeline for fibroblast-to-neuron reprogramming with ATAC-seq, ChIP, ChIP-seq, and RNA-seq experiments annotated. DPI, days post infection; DPT, days post treatment with FD.
(B) Diagram representing the effects of FD exposure on NEUROG2 peak score for binding events detected at 0.5, 1, and 2 DPT. The change in peak intensity is represented as a ratio of the peak score in the presence versus absence of FD with the Venn diagram depicting the number of NFD-unique events at 0.5, 1, and 2 DPT (biological replicates, n = 3 for each time point and condition).
(C) Hierarchical clustering and heatmap of changes in transcription detected by RNA-seq relative to control fibroblasts. FD, fibroblasts exposed to FD; N, NEUROG2-transduced fibroblasts; NFD, NEUROG2-transduced fibroblasts exposed to FD (biological replicates, n = 3 for each time point and condition).
(D) Pie diagrams representing the respective number of genes induced by NEUROG2, FD, and NEUROG2-FD synergistic action at 2 DPT. Synergy represents genes only induced in the NFD condition.
(E) Time-course representation of the change in transcript expression intensity for genes regulated by FD, NEUROG2, or NFD synergy.
(F) Sunburst diagram depicting the upregulated and downregulated genes at each time point, the intensity of expression change at 2 DPT, and corresponding NEUROG2 ChIP-seq annotation data. Each segment represents one collection of time points. For example, the section with pink, red, and deep-red segments represents genes with significant differential expression at all time points, while the section with only a pink segment represents genes differentially expressed only at 0.5 DPT. The gray outer gradient represents the expression change observed for the genes in each section. NEUROG2 binding events are annotated with a blue line segment.