Figure 3.
Small Molecules Activate CREB1 to Promote Neuron Survival and SOX4 Upregulation
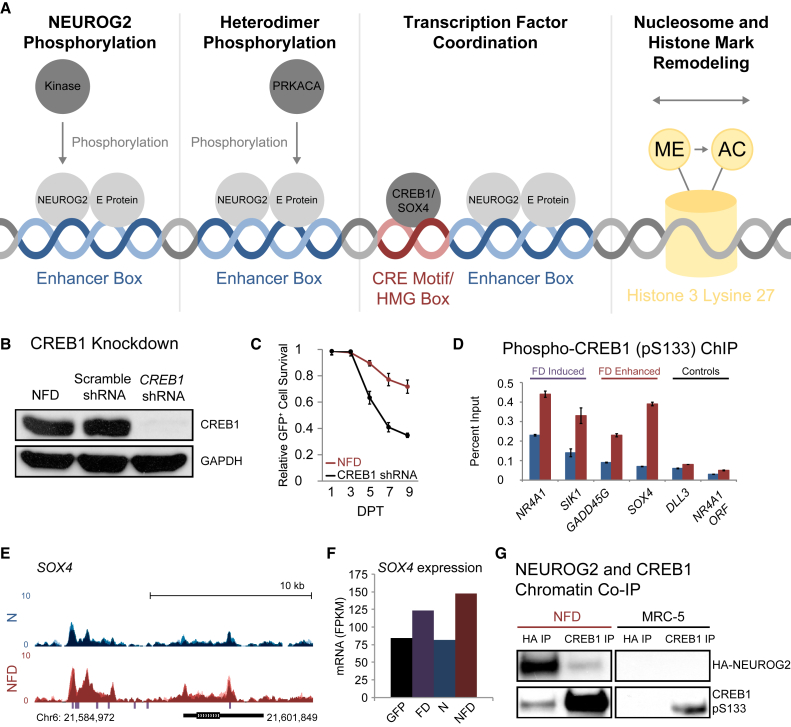
(A) Four hypothetical mechanisms that might underlie NEUROG2 and FD synergy.
(B) Immunoblot of shRNA-mediated CREB1 knockdown.
(C) CREB1 knockdown significantly reduces the viability of reprogrammed neurons (mean ± SD; n = 10 random image fields from biological triplicates). Relative survival represents (GFP+ and Hoechst 33342+ cells at each time point)/(total GFP+ and Hoechst 33342+ cells at 1 DPT).
(D) Phospho-CREB1 (pS133) ChIP-qPCR at CREB1 targets (NR4A1, SIK1), NEUROG2 targets with FD-enhanced expression (GADD45G, SOX4), and controls (NEUROG2 target, DLL3; within gene, NR4A1 open reading frame) for NEUROG2-transduced fibroblasts (blue) and NFD-treated fibroblasts (red) at 1 DPT (mean ± SD; biological triplicates).
(E) NEUROG2 occupancy around the SOX4 locus. NEUROG2 occupancy is represented as an overlay of 0.5-DPT (light), 1-DPT (medium), and 2 DPT (dark) NEUROG2 ChIP-seq traces from NEUROG2-transduced fibroblasts (blue) and NFD-treated fibroblasts (red). Purple hashes indicate CRE motif sequences.
(F) Condition-dependent SOX4 expression from RNA-seq results at 2 DPT (mean values from biological triplicate RNA-seq data).
(G) NEUROG2 and CREB1 ChIP from control and NFD-treated fibroblasts reveals chromatin co-occupancy in NFD-treated fibroblasts.