Abstract
Identification of epigenetic reversal agents for use in combination chemotherapies to treat human pancreatic ductal adenocarcinomas (PDAC) remains an unmet clinical need. Pharmacological inhibitors of Enhancer of Zeste Homolog 2 (EZH2) are emerging as potential histone methylation reversal agents for the treatment of various solid tumors and leukemia; however, the surprisingly small set of mRNA targets identified with EZH2 knockdown suggests novel mechanisms contribute to their anti-tumorigenic effects. Here, 3-deazaneplanocin-A (DZNep), an inhibitor of S-adenosyl-L-homocysteine hydrolase and EZH2 histone lysine-N-methyltransferase, significantly reprograms noncoding miRNA (miR) expression and dampens TGFβ1-induced epithelial-to-mesenchymal (EMT) signals in pancreatic cancer. In particular, miR-663a and miR-4787-5p were identified as PDAC-downregulated miRs that were reactivated by DZNep to directly target TGFβ1 for RNA interference. Lentiviral overexpression of miR-663a and miR-4787-5p reduced TGFβ1 synthesis and secretion in PDAC cells and partially phenocopied DZNep’s EMT-resisting effects, whereas locked nucleic acid (LNA) antagomiRs counteracted them. DZNep, miR-663a, and miR-4787-5p reduced tumor burden in vivo and metastases in an orthotopic mouse pancreatic tumor model. Taken together, these findings suggest the epigenetic reprogramming of miRs by synthetic histone methylation reversal agents as a viable approach to attenuate TGFβ1-induced EMT features in human PDAC and uncover putative miR targets involved in the process.
Keywords: DZNep, EMT, TGF-β1, Pancreatic Cancer, miR-663a, miR-4787-5p
INTRODUCTION
The tendency of pancreatic tumors to metastasize to lymph nodes, the abdominal cavity, and the liver at a very early stage compromises the effectiveness of chemotherapeutic agents (1, 2). Hence, challenges to prevent tumor spread and drug resistance continue to exist for pancreatic cancer treatment. Evidence shows EMT, distinct cellular events characterized by loss of epithelial phenotypes and gain of mesenchymal phenotypes, contributes to the early dissemination of primary tumor cells and metastatic spread (2–4). Further, the process of EMT itself has been shown to promote stem-like characteristics of tumor cells and increase drug resistance (2–4). Therefore, therapeutic strategies directed against EMT would likely prove useful in controlling metastasis and drug resistance in PDAC patients.
Transforming growth factor beta 1 (TGF-β1) is one of the critical cytokines known to drive EMT in pancreatic cancer (5, 6). It binds and activates TGF-β Receptor 2 (TGFBR2), propagating signaling through phosphorylation of Smad proteins and associating with transcription factors to regulate gene expression (7). Although the primary roles of TGF-β1 are to inhibit epithelial cell proliferation and orchestrate mesenchymal development (8), loss of growth control responses and activation of EMT become salient in advanced tumors (9, 10). These outcomes are due to the loss of normal responsiveness to the ligand as a result of either mutations in TGF-β receptors (e.g., TGFBR2) and/or intracellular SMAD proteins (e.g., Smad4) (11, 12). Other non-genetic mechanisms have also shown to contribute to aberrant TGF-β signaling (13, 14). Evidence also presents that the aforementioned events, largely in a stochastic manner, allows transcriptional reprogramming of a network of EMT-related genes and alterations in the nucleosome state to promote a sustained mesenchymal signature (9, 15).
Recent studies have uncovered novel epigenetic mechanisms such as methylation of lysine in histones to preferentially amplify pro-tumorigenic signaling of cancer cells to EMT (16–19). In particular, a striking correlation between TGF-β1 and EZH2, a histone methyl transferase enzyme in the Polycomb Repressive Complex 2 (PRC2) involved in silencing of genes via H3K27Me, was recognized to regulate EMT (20, 21). Other members of the PRC2 including EED and JARID2 have also been reported to drive TGF-β-induced EMT, suggesting a possible widespread effect of histone methylation in the steps leading to EMT (22, 23). Unlike the advancements in the biological understanding of EZH2 on TGF-β-induced EMT, the cancer pharmacology of EZH2 inhibitors is lagging behind. Despite new and more specific EZH2 inhibitor candidates currently being evaluated in clinical trials for lymphoma and multiple myeloma (24), their utilities in metastatic control of solid tumors remains obscure. We have previously reported DZNep, a carbocyclic analog of adenosine that depletes cellular levels of EZH2 and erases the H3K27Me mark, to curtail growth, proliferation, and anticancer nucleoside analog resistance of pancreatic cancer cells (25). Here, we report DZNep’s role in dampening TGF-β1-induced EMT features in PDAC. We present evidence for DZNep-induced miRNA reprogramming as a mechanism for EMT inhibition in PDAC.
MATERIALS & METHODS
Cell Culture, MTT Cytotoxicity Assays, Clonogenic Assays, Western Blotting, Real-time PCR Analysis, and MiRNA Overexpression
All cell lines were received from ATCC (Manassas, VA) and were used within 25 passages, not exceeding a period of 2–3 months of revival. The ATCC uses morphological, cytogenetic and DNA profile analyses for characterization of cell lines. Human pancreatic ductal epithelial (HPDE) cells were received from Dr. Ming Tsao of the Ontario Cancer Institute (Toronto, Canada). The L3.6pl cell line was received from Dr. Isiah D. Fidler at The University of Texas MD Anderson Cancer Center (Houston, TX). Both HPDE and L3.6pl cell lines were handled as other cell lines and were genotyped by DNA fingerprinting (PowerPlex 16; Promega, Madison, WI) as per the manufacturer’s instructions. The growth conditions of cell lines were performed as described previously (26). Panc 10.05 was grown in RPMI-1640 Medium with 15% FBS and 10 U/ml human recombinant insulin while CFPAC-1 was grown in Iscove’s Modified Dulbecco’s Medium with 10% FBS.
Tumor RNA
Total RNA from normal pancreas and PDAC were procured from Asterand (Detroit, MI) (Table S1).
Materials, Scratch Wound Assay, Transwell Invasion Assay, ELISA, MicroRNA Microarray, Luciferase In Vitro Reporter Assay, Knockdown of miRNA, RT2 Profiler PCR Array, Orthotopic Pancreatic Tumor Xenograft Model (27), and Statistical Analysis
RESULTS
DZNep resists TGF-β1-induced EMT in pancreatic cancer cells
To investigate DZNep effects on TGF-β1-induced EMT, we tested TGF-β1-induced changes in morphology and growth of two moderately-poorly differentiated PDAC cell lines, viz. MIA PaCa-2 and PANC-1. Recombinant-derived human TGF-β1 (10 ng/ml; 72 h) induced distinct EMT-like, morphological changes in both MIA PaCa-2 and PANC-1 (Fig. 1A) but not in normal HPDE (data not shown). More spindle shaped cells with elongated cellular processes and diminished cell-to-cell contacts (Fig. 1A) as well as reduced expression of epithelial markers (E-cadherin and cytokeratin8/18) and increased expression of mesenchymal markers (N-cadherin and vimentin) were noted with TGF-β1 treatment (Fig. 1B). TGF-β1-induced EMT changes were independent of changes in cell proliferation in MIA PaCa-2 with only a slight growth reduction in PANC-1 (12.23±0.35%; p<0.05; Fig. S1–S2). On the contrary, TGF-β1 significantly reduced cell proliferation in normal HPDE (30.25±1.99%; p<0.005; Fig. S1). These data confirmed the presence of TGF-β1-mediated EMT-like features in MIA PaCa-2 and PANC-1 and were therefore used for further studies.
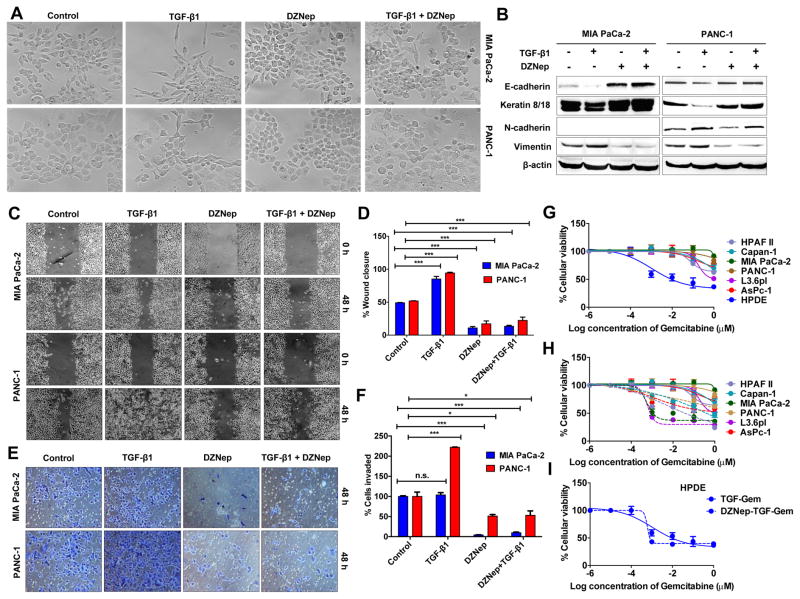
Figure 1. DZNep inhibits EMT and chemoresistance in pancreatic cancer.
A. DZNep resisted TGF-β1-induced morphological EMT features. Phase contrast images of live cells after treatments (72 h). Original magnification, X10. B. DZNep resisted TGF-β1-induced changes in epithelial and mesenchymal markers. Whole cell lysates (50 μg) from cells treated with TGF-β1, DZNep, or both for 72 h subjected to Western blotting for EMT markers. β-actin, the internal loading control, is shown with a representative blot. The position of a non-specific band is indicated by an asterisk (*). C. Representative images of cell monolayers subjected to a scratch wound assay shows DZNep inhibited cell migration. Original magnification X4. D. Quantification of wound closure measurements. E. A representation of cells invaded into a Matrigel-coated transwell insert after crystal violet staining. F. Invaded cells were counted and plotted. G & H. DZNep resisted TGF-β1-induced gemcitabine chemoresistance in pancreatic cancer cell lines. 3X103 cells seeded in a 96-well plate were treated with TGF-β1 (24 h) in the presence (G; dotted lines) or absence (G & H; solid lines) of DZNep (24 h) followed by an MTT cytotoxicity analysis with gemcitabine. I. DZNep did not increase cytotoxicity in TGF-β1 treated HPDE. For all experiments, cells were treated with DZNep at 10 μM and TGF-β1 at 10 ng/mL. Points, mean of triplicates; bars, SD. n=3.
In sub-confluent conditions, MIA PaCa-2 appears more plastic with the presence of both epithelial- and mesenchymal-looking cells while PANC-1 appears predominantly epithelial (Fig. 1A). Interestingly, DZNep treatment visibly increased the epithelial morphological characteristics of both cell lines that were more readily identifiable in MIA PaCa-2 than PANC-1 (Fig. 1A). Biochemically, DZNep increased expression of E-cadherin and cytokeratin-8/18 and decreased expression of vimentin and N-cadherin in PANC-1 as judged by Western blotting analysis (Fig. 1B). Similar results were also obtained in MIA PaCa-2 (except that N-cadherin was not expressed; Fig. 1B), L3.6pl, PANC10.05, CFPAC-1, and SW1990 (Fig. S3) suggesting the effects are widespread in PDAC. DZNep treatment also resisted migratory and invasive characteristics of MIA PaCa-2 and PANC-1 (Fig. 1C–1F) and even retained its growth inhibitory effects in the presence of TGF-β1 (Fig. S2). Combined treatment of DZNep and TGF-β1 opposed the morphological and biochemical EMT changes in MIA PaCa-2 and PANC-1 (Fig. 1A–1B; Fig S4). Further, the presence of DZNep significantly inhibited TGF-β1-induced increases in cell migration and invasion in MIA PaCa-2 and PANC-1 (Fig 1C–1F). DZNep in the presence of TGF-β1 increased gemcitabine cytotoxicity in PDAC cell lines (Fig. 1G–1H) but not in HPDE (Fig. 1G & 1I). Together, these data identified DZNep’s antagonistic effects on TGF-β1-induced EMT features in PDAC cell lines.
DZNep inhibits endogenous TGF-β1 protein expression and secretion in a dose- and time-dependent manner
The increase in epithelial characteristics of inherently mesenchymal MIA PaCa-2 (Fig. 1A; top panel labeled DZNep) was suggestive of the inhibition of endogenous TGF-β1 by DZNep. To test this possibility, we assayed the endogenous TGF-β1 in MIA PaCa-2 and PANC-1. Since cellular TGF-β1 exists as either a latent TGF-β1 complex (290 kDa) or a biologically active monomeric or homodimeric form (13 kDa and 26 kDa, respectively) (28), we first examined the identity of TGF-β1 in MIA PaCa-2 and PANC-1. Western blotting analysis displayed endogenous TGF-β1 in both cell lines to migrate predominantly as a 26 kDa homodimeric TGF-β1, although a band corresponding to the monomeric form (13 kDa) was faintly detectable in some cases (Fig. 2A). A non-specific ~20 kDa protein was also detectable. Under the same conditions, recombinant hTGF-β1 also migrated as both monomeric and dimeric forms (Fig. 2A), so we focused on the better-detectable 26 kDa homodimeric bands for subsequent experiments. Treatment with DZNep distinctly reduced the 26 kDa TGF-β1 immunoreactivity in both MIA PaCa-2 and PANC-1 in a dose- and time-dependent fashion (Fig. 2A). Significant reductions in TGF-β1 were evident from 48 h of 10 μM DZNep treatment with almost complete loss of immunoreactivity observed at ~96–120 h in MIA PaCa-2 (Fig. 2A). Reduction in TGF-β1 with DZNep treatment was also evident in several other PDAC cell lines (Fig. 2B). Intriguingly, none of these changes in TGF-β1 expression appeared to be transcriptionally regulated as real time PCR analysis of total RNA extracted from DZNep-treated cells showed a lack of change in TGF-β1 transcripts (Fig. 2C & S5).
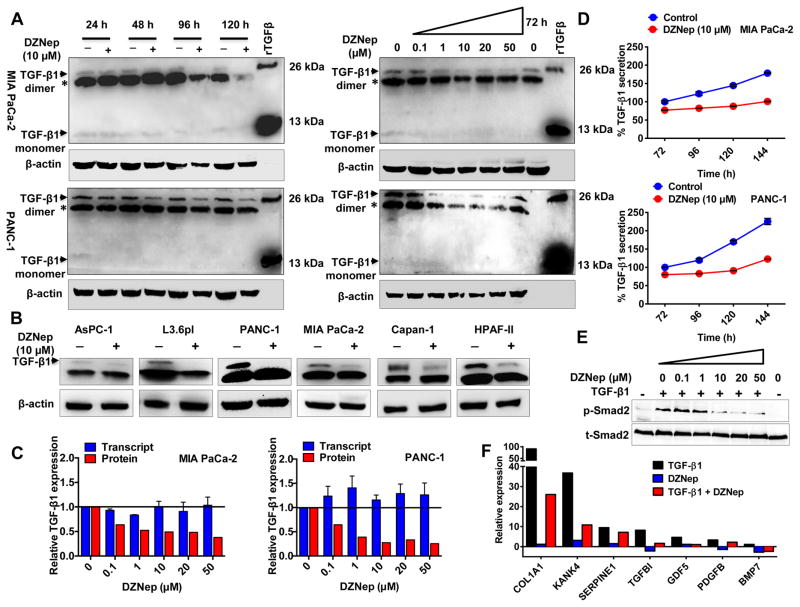
Figure 2. DZNep inhibits TGF-β1 synthesis and secretion in pancreatic cancer.
A. DZNep decreased TGF-β1 protein in a time- and dose-dependent manner. Whole cell lysates (50 μg) of DZNep-treated cells subjected to Western blotting for TGF-β1 expression. A rhTGF-β1 used as a positive control and β-actin as a loading control. B. DZNep decreased TGF-β1 protein in PDAC cell lines. C. DZNep mediated decrease in TGF-β1 was independent of transcriptional changes. Densitometric values of TGF-β1 protein (A, right) and transcript levels were compared. GUSB used as an internal control for qRT-PCR. D. DZNep reduced TGF-β1 secretion into the culture supernatants. The extent of decrease in secretion was measured by ELISA. E. DZNep attenuated TGF-β1 signaling in a dose-dependent fashion. Whole cell lysates of PANC-1 treated with TGF-β1 (10 ng/mL; 60 mins) and DZNep (24 h pre-treatment) subjected to Western blotting for total and p-Smad2. F. DZNep decreased transcript levels of TGF-β-responsive genes as determined by qRT-PCR. Points, mean of triplicate; bars, SD. n=3.
Since TGF-β1 plays a vital role in modulating tumor microenvironment via autocrine/paracrine mechanisms (29), we examined whether DZNep can inhibit secretory levels of TGF-β1. Using a quantitative sandwich ELISA, we assayed the culture media of DZNep-treated PDAC cells for changes in extracellular TGF-β1 (Fig. 2D). Consistent with a reduction in TGF-β1 protein, DZNep (10 μM) decreased TGF-β1 secretion into the extracellular media in a time-dependent manner (~40–50% reduction in 5–6 days) (Fig. 2D). In addition, we tested whether the reductions in TGF-β1 protein and secretion can affect its downstream signaling by investigating the phosphorylation status of Smad2 (a critical mediator of TGF-β signaling pathway) in DZNep-treated PANC-1 (Fig. 2E). Western blotting analysis revealed DZNep to significantly reduce p-Smad2 without altering total Smad2 (Fig. 2E). Consistently, DZNep significantly inhibited the expression of several known TGF-β1-induced genes (Fig. 1F). Collectively, these data identified that DZNep can significantly inhibit TGF-β1 synthesis and secretion as well as TGF-β signaling in PDAC.
DZNep-induced epigenetic reprogramming of microRNAs in pancreatic cancer cells
The mechanism of DZNep inhibition of TGF-β1 remained unclear, but it was unlikely that TGF-β1 was regulated at the transcriptional level. The lack of a major change in the slope of the line representing TGF-β1 secretory levels after DZNep treatment (compared with untreated control) (Fig. 2D) also argued against probable alterations in the post-translational stability of TGF-β1. Hence, we postulated that DZNep could reprogram miRNAs in cancer cells to repress TGF-β1 protein by RNA interference. To test this hypothesis, we screened >1900 human miRNAs (referenced in the Sanger miRBase Release 18.0.) using microarrays to identify possible global alterations in MIA PaCa-2 treated with DZNep as opposed to untreated MIA PaCa-2 (Fig. 3A & 3B). Further, the miRNA profile of MIA PaCa-2 was compared with that of HPDE to understand miRNAs differentially expressed (Fig. 3C & S6). The heat map generated from hierarchical cluster analysis showed a significant reduction in miRNAs in cancer cells with 52.3% of miRNAs (91 out of 174 detectable miRNAs) downregulated in MIA PaCa-2 (Fig. 3C–3E & S6; Table S3). Similarly, we identified 68.8% of miRNAs (161 out of 234 detectable miRNAs) downregulated in 10 different patient-derived PDAC tissues compared with two normal pancreatic tissues (data not shown). Thirty-five miRNAs were found commonly downregulated in both MIA PaCa-2 and PDAC tissues (Table S4). Interestingly, DZNep increased the expression of >50 miRNAs (22 miRNAs at 8 h; 34 miRNAs at 72 h) (Fig. 3B & 3D; Table S5) and even partially reversed the downregulation pattern of miRNAs seen in MIA PaCa-2 (Fig. 3E & 3F; Table S6).
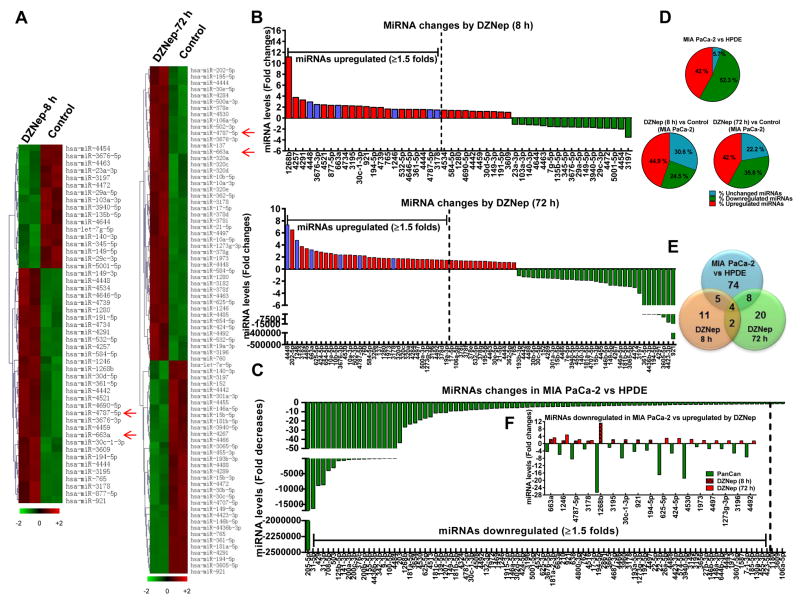
Figure 3. DZNep reprograms PDAC-downregulated miRNAs in pancreatic cancer.
A. Clustering of miRNAs differentially expressed in DZNep (1 μM)-treated MIA PaCa-2 versus untreated. Heat maps with statistically significant (p<0.1) changes in miRNA expression shown by a 2-color system (light green, lowest expression; dark red, highest expression). Each row represents expression of a single miRNA while each column represents a single sample. n=2. B. Fold changes in miRNAs after 8 h or 72 h of DZNep treatment in MIA PaCa-2. Purple bars indicate miRNAs upregulated by DZNep at both time points. C. MiRNAs downregulated in MIA PaCa-2 compared against HPDE. D. Pie charts showing expression patterns of miRNAs. A fold change of ≥ or ≤1.5 considered as upregulated or downregulated respectively. E. Venn diagram comparing miRNAs downregulated in MIA PaCa-2 against HPDE (blue; n=2) with those upregulated by DZNep in MIA PaCa-2 at 8 h (orange) or 72 h (green). F. Fold changes of miRNAs downregulated in MIA PaCa-2 (Pan Can) but upregulated after DZNep treatment.
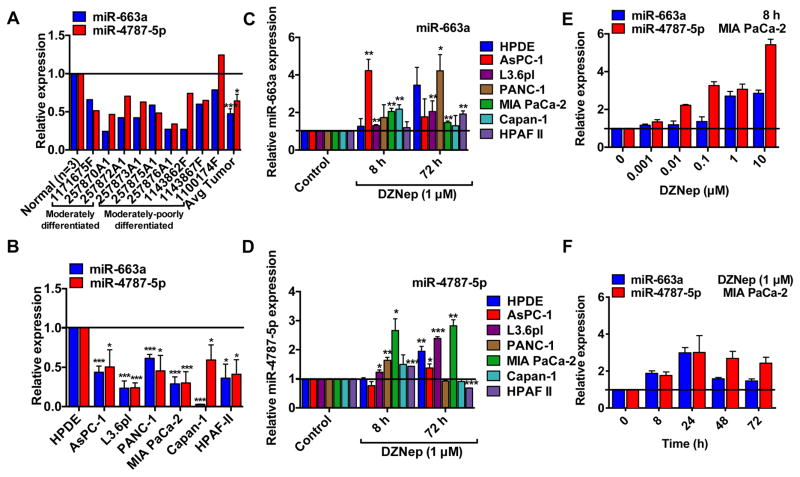
We next examined whether any of the DZNep-upregulated miRNAs can target TGF-β1 for RNA interference. First, we utilized publicly available algorithms to predict binding of DZNep-upregulated miRNAs to the 3′UTR of TGF-β1. The initial analysis identified several DZNep-upregulated miRNAs to possibly regulate known players in the TGF-β signaling pathway including TGF-β1 (Table S8). MiR-663a and miR-4787-5p in particular were predicted to bind to the 3′UTR of TGF-β1 (Table S8). Interestingly, those miRNAs were significantly downregulated by 5–10 folds in both MIA PaCa-2 (Fig. 3F). Further comparison of DZNep upregulated miRNAs with those downregulated in MIA PaCa-2 showed that DZNep restored expression of 17 miRs with miR-663a and miR-4787-5p expressions restored at both the 8 h and 72 h time points (Fig. 3E & 3F). Real-time PCR analysis with individual primers/probes further validated downregulation of miR-663a and miR-4787-5p in several PDAC tissues (Fig. 4A) and cell lines (Fig. 4B) as well as restoration of expression with DZNep in PDAC cell lines (Fig. 4C & 4D). DZNep-induced expression of miR-663a and miR-4787-5p was dose- and time-dependent with maximal changes observed within 8–24 h of 0.1–1 μM DZNep treatment in MIA PaCa-2 and PANC-1 (Fig. 4E & 4F). Induction of miR-663a and miR-4787-5p was specific to DZNep and no other clinically-used nucleoside analogs (Fig. S7). Importantly, EZH2 shRNA induced miR-663a and miR-4787-5p levels and decreased TGF-β1 expression in MIA PaCa-2 cells suggesting EZH2 silencing can phenocopy DZNep’s actions (Fig. S8).
Figure 4. DZNep restores miR-663a and miR-4787-5p expression in pancreatic cancer.
A & B. MiR-663a and miR-4787-5p are downregulated in patient-isolated PDAC tissues (A) and cell lines (B) as determined by qRT-PCR. GUSB and U6B were used as an internal control for assaying miR-663a and miR-4787-5p. C & D. DZNep increased miR-663a (C) and miR-4787-5p (D) expression in PDAC cell lines. E and F. DZNep increased miR-663a and miR-4787-5p expressions in a dose- and time-dependent manner. Bars, SD. n=3.
DZNep-induced miR-663a and miR-4787-5p directly target TGF-β1 for RNA interference
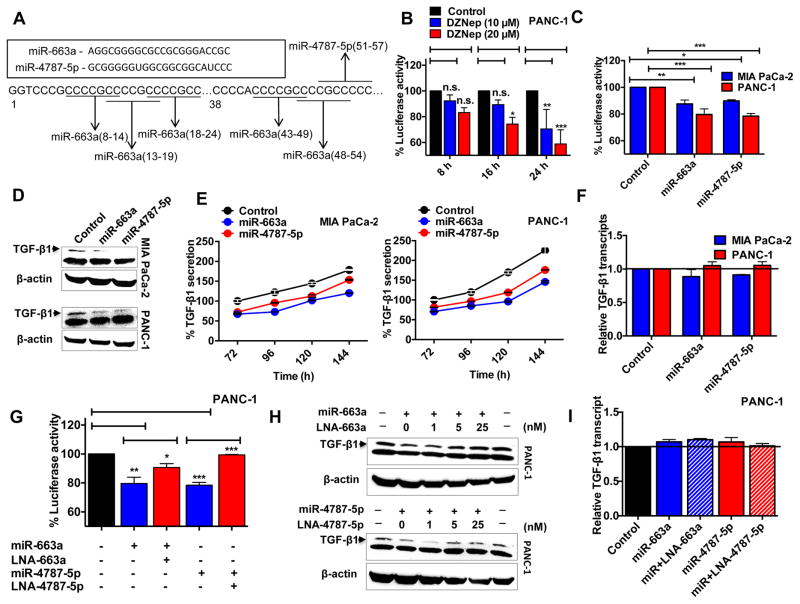
On subsequent DNA analysis, we identified five binding sites for the miR-663a seed sequence and a single probable binding site for miR-4787-5p within a 60 bp region of the beginning of the TGF-β1 3′UTR (Fig. 5A). To experimentally evaluate whether miR-663a or -4787-5p can target TGF-β1 for RNA interference, we examined the changes in the expression of an engineered dual luciferase construct in which the 3′UTR of TGF-β1 was cloned downstream of a luciferase cDNA. Interestingly, DZNep treatment of PANC-1 itself significantly destabilized the luciferase activity of the engineered construct in both a dose- and time-dependent fashion (Fig. 5B), providing preliminary evidence for the DZNep reprogrammed miRNAs to alter binding interactions with TGF-β1 3′UTR. To test if miR-663a or miR-4787-5p can directly bind to the TGF-β1 3′UTR and direct the transcripts for RNA interference, we repeated the luciferase assays on MIA PaCa-2 and PANC-1 stably expressing miR-663a or miR-4787-5p (Fig. 5C). Stable expression was achieved using lentiviral gene constructs wherein ~3–5-fold and ~10–12-fold increases in miR-663a and miR-4787-5p expression, respectively, was achieved (Fig. S9). The results obtained from these luciferase assays indicated that both miR-663a and miR-4787-5p can directly bind to the 3′UTR of TGF-β1 and destabilize luciferase expression in MIA PaCa-2 and PANC-1 (Fig. 5C). Western blotting analysis using cell lysates and ELISA on culture supernatant showed a reduction in TGF-β1 protein synthesis (Fig. 5D) and secretion (Fig. 5E) in cells overexpressing miR-663a and miR-4787-5p. Again, these changes were independent of TGF-β1 transcriptional variations (Fig. 5F). The reductions in luciferase activities noted with miRNAs were effectively counteracted with their respective LNA-derived antagomirs (Fig. 5G), suggesting the observed effects were specific to miR-663a and miR-4787-5p. Additionally, LNA-derived antagomiRs significantly antagonized the respective miRNA-induced reductions in TGF-β1 protein (Fig. 5H–5I). These data identified the direct binding of miR-663a and miR-4575-5p to the 3′UTR of TGF-β1 and supported RNA interference as a mechanism of DZNep inhibition of the TGF-β signaling pathway in PDAC.
Figure 5. MiR-663a and miR-4787-5p directly targets TGF-β1 for RNA interference.
A. Schematic representation of predicted miR-663a and miR-4787-5p binding site(s) in the TGF-β1 3′-UTR. B. DZNep reduced luciferase activity in cells transfected with the TGF-β1 3′-UTR luciferase reporter construct. C. MiR-663a or miR-4787-5p directly binds to TGF-β1 3′UTR. Cells stably overexpressing miRNAs were transfected with the TGF-β1 3′-UTR luciferase reporter and percent activities recorded. D. MiR-663a and miR-4787-5p decreased TGF-β1 protein. Whole cell lysates (50 μg) of cells stably overexpressing miRNAs subjected to Western blotting to determine TGF-β1 protein levels with β-actin as a loading control. E. MiRNA overexpression decreased TGF-β1 secretion into the culture supernatants. F. MiR-663a and miR-4787-5p did not affect TGF-β1 transcripts. Total RNA from cells stably overexpressing miRNAs subjected to qRT-PCR to determine TGF-β1 transcript levels with GUSB as an internal control. G. LNA-663a or LNA-4787-5p (50 nM; 48 h) antagonized respective miRNA-induced decreases in luciferase activity of cells. H & I. LNA-663a or LNA-4787-5p antagonized respective miRNA-induced decreases in TGF-β1 protein levels (H) but had no effect on TGF-β1 transcripts (at 50 nM; 48 h) (I). Points, mean of triplicate; bars, SD. n=3.
miR-663a and miR-4787-5p contribute to DZNep-mediated EMT control in pancreatic cancer
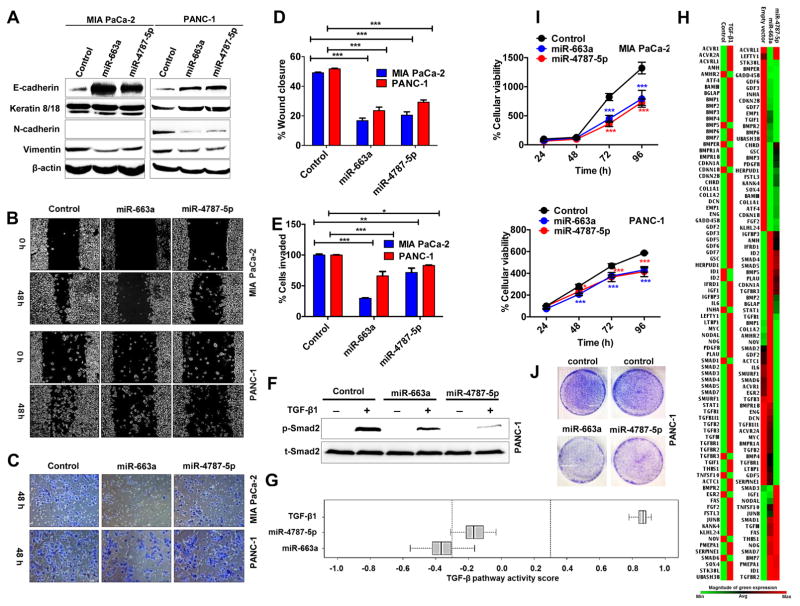
We next investigated whether miR-663a and miR-4787-5p can suppress TGF-β1-induced EMT in PDAC as observed with DZNep (Fig. 1). Overexpression of either miR-663a or miR-4787-5p significantly induced E-cadherin and decreased vimentin (Fig. 6A) similar to DZNep (Fig. 1B). Both miR-663a and miR-4787-5p also increased cytokeratin 8/18 and decreased N-cadherin in PANC-1 (Fig. 6A). Both miR-663a and miR-4787-5p overexpressing cells exhibited reduced cell migration (Fig. 6B & 6D) and invasion (Fig. 6C, 6E & S10) similar to that observed with DZNep treatment. Morphologically, miR-663a and miR-4787 overexpression visibly increased the epithelial appearance of MIA PaCa-2 as observed with DZNep treatment; however, the changes between control and miR-overexpressing PANC-1 were indistinguishable (data not shown). Both miR-663a and -4787-5p significantly diminished the p-Smad2 level without altering total Smad2 in PANC-1 (Fig. 6F). As seen with DZNep, miR-663a and miR-4787-5p also significantly reduced expression of the TGF-β responsive genes examined in Fig. 1F. However, to further extend the understanding of the miR-induced effects on the entire TGF-β axis, we profiled transcript changes of 84 pathway-related genes (Table S2). PCR array data analyses of Ct values identified a negative TGF-β pathway scores of −0.4 for miR-663a (p<0.01) and −0.2 for miR-4787-5p (p=n.s.), respectively, confirming the repressive roles of miRNAs on TGF-β signaling pathway (Fig. 6G & 6H). As expected, TGF-β activated the pathway and generated a positive pathway score of +0.865 (p<0.001; Fig. 6G & 6H). In addition to attenuation of TGF-β signaling, both miRNAs moderately reduced growth and cell proliferation rates in MIA PaCa-2 and PANC-1 cells as observed from MTT and colony formation assays (Fig. 6I & 6J).
Figure 6. MiR-663a and miR-4787-5p resists EMT features and slows cellular growth in pancreatic cancer cells.
A. MiR-663a and miR-4787-5p increased epithelial markers and decreased mesenchymal markers. Whole cell lysates (50 μg) of cells overexpressing miRNAs were subjected to Western blotting for EMT markers with β-actin as a loading control. B & D. MiR-663a and miR-4787-5p decreased cell migration. Representative images of cells subjected to a scratch wound assay (B) and quantification of wound closure measurements (D). Original magnification X4. C & E. MiR-663a and miR-4787-5p decreased cell invasion. F. MiR-663a and miR-4787-5p reduced p-Smad2 levels. Whole cells lysates of TGF-β1 (10 ng/mL; 60 minutes) treated cells subjected to Western blotting to detect total and p-Smad2. G & H. MiRNAs decreased expression of TGF-β1-responsive genes and attenuated TGF-β1 signaling pathway. Total RNA from cells subjected to a RT2-PCR profiler array. Heat maps showing transcript level changes of TGF-β-responsive genes (H) and overall TGF-β pathway activity scores depicted (G). I & J. MiR-663a and miR-4787-5p reduced cellular proliferation as determined by MTT (I) and colony formation (J) assays. Points, mean of triplicate; bars, SD. n=3
DZNep, miR-663a, and miR-4787-5p suppress the growth and metastasis of pancreatic cancer xenografts
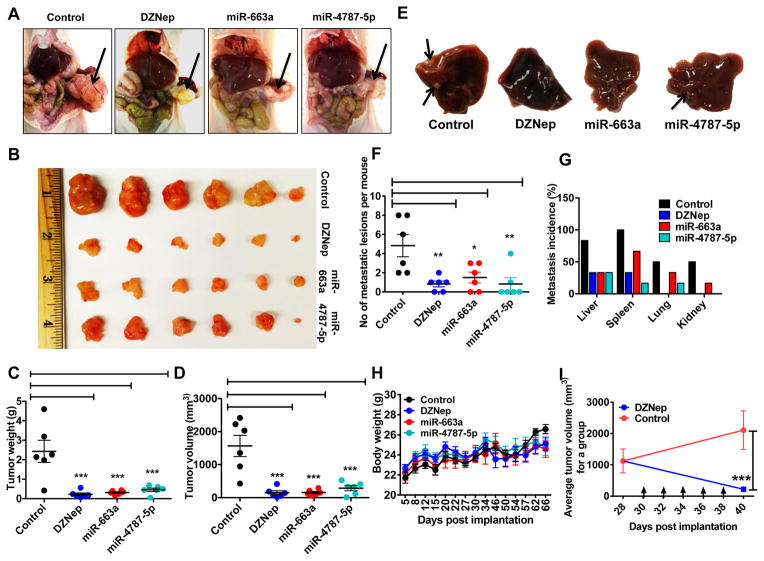
Finally, we investigated whether the effects of DZNep and miRNAs on EMT resistance could be recapitulated in vivo. Athymic nude mice were orthotopically implanted with PANC-1 stably transduced with control, miR-663a, or miR-4787-5p lentiviral constructs. Mice injected with control PANC-1 were also treated with DZNep (3 mg/kg, IP, twice a week) until sacrificed. At the end of 10 weeks, tumor growth was found to be significantly smaller in DZNep-treated mice as well as in mice transplanted with miR-663a or miR-4787-5p cells as compared to untreated mice injected with control cells (Fig. 7A & 7B). Consistently, 10-week tumor weights (Fig. 7C) and volumes (Fig. 7D) were significantly less for DZNep-treated group as compared with control group. Mice transplanted miR-663a or miR-4787-5p overexpressing cells also showed reduced tumor weights and volumes against control groups (Fig. 7A–D). Furthermore, as predicted from in vitro data (Fig. 1 & Fig. 6), the total number of metastatic lesions in the secondary organs (liver, spleen, lungs, and kidneys) were significantly less in DZNep-treated (mean=0.83±0.8), miR-663a-tranduced (mean=1.5±1.4), and miR-4787-5p-transduced (mean=0.83±1.6) mice as compared to control groups (mean=4.83±2.9) (Fig. 7E–G). In fact, 67% of mice in the DZNep-treated or miRNA-transduced group showed no metastatic lesions in the liver (main site of pancreatic cancer metastasis) while 83.33% of mice in the control group demonstrated visible liver metastases. DZNep-treated or miRNA-transduced mice also exhibited reduced metastatic foci in the spleen, lungs, and kidneys as compared to control groups (Fig. 7F & 7G). Overall, DZNep and miRNAs in tumors were well tolerated by the mice as evident by no significant changes in hepatic enzyme (sGPT and sGOT) levels or body weight (Fig. 7H). Finally, DZNep also induced regression of tumors in mice (Fig. 7I & Fig. S11). These results support that DZNep and miR-663a and miR-4787-5p can suppress the metastatic capacity of orthotopically implanted pancreatic tumor cells.
Figure 7. DZNep and miRNAs suppress pancreatic cancer growth and metastasis in vivo.
A & B. DZNep and miRNAs reduced primary tumor burden in an orthotopic pancreatic xenograft model. Representative photographs of tumor xenografts (A) and excised tumor specimens (B) after 10 weeks of cell injections. C & D. Average tumor weights (C) and volumes (D) in DZNep treated and miRNA-expressed conditions. Each point represents data obtained from one mouse within a group. E. Representative images of tumor metastases to the liver with arrows pointing to metastatic foci. Tumor characteristics were confirmed by histopathology. F & G. DZNep, miR-663a, and miR-4787-5p reduced overall metastasis in secondary organs. F. The total number of metastatic nodules per mouse was added and plotted as a data point. The bar indicates mean number of lesions per mouse within a group. G. Metastatic incidences per organ. H. Average body weights of mice with each point representing average mice weight (n=6) for a group. I. DZNep induced regression of tumors in mice (n=3 for each group). Arrows indicate days of DZNep treatment. Bars, SD.
Discussion
Increasing evidence is recognizing the critical role of EZH2-induced chromatin alterations and transcriptional reprogramming of genes in EMT (20). For instance, SOX4 was identified as a master regulator of EMT by directly controlling EZH2 expression in normal and cancerous breast epithelial cells (21). EZH2 was shown to support cancer cell invasion and metastasis via the TGF-β1 axis in various solid tumors and serves as a predictive indicator of treatment outcomes in PDAC and ovarian cancer patients (30, 31). Despite recognizing EZH2 regulation of EMT, studies on the development of pharmacological inhibitors of EZH2 to mitigate EMT progression are limited. While DZNep inhibition of EZH2 has been shown to increase the expression of the epithelial marker E-cadherin in certain cancer types (e.g., renal cell carcinoma) (32), its role in modulating overall EMT characteristics in solid tumors remains elusive. The current study uncovers the role of DZNep in dampening TGF-β1-induced EMT characteristics in PDAC. To our knowledge, this is the first study that demonstrates a direct effect of a histone methyltransferase inhibitor in decreasing TGF-β1 protein and secretory levels in cell lines representing various subtypes of PDAC. Pharmacological treatment with DZNep distinctly induced epithelial characteristics of PDAC cells, counteracted TGF-β1-induced mesenchymal characteristics, and inhibited migratory and invasive signals that drive tumor progression. The mechanism of DZNep inhibition of TGF-β1 was identified as epigenetic reprogramming of miRNAs, which included a subset of miRNAs downregulated in PDAC. Significant contributions came from two epigenetically reprogrammed miRNAs (miR-663a and miR-4787-5p) that directly targeted TGF-β1 for RNA interference and reduced TGF-β1 synthesis and secretory levels. These findings reveal an alternative mechanism of TGF-β1 regulation of EMT in PDAC and better explain the anti-metastatic effects of synthetic histone methyl transferase inhibitors that have so far only been known to upregulate a small number of coding genes.
Earlier we reported DZNep to induce delayed cytotoxic and apoptotic effects in moderately-poorly differentiated MIA PaCa-2 and PANC-1 (25). While we demonstrate here that DZNep can also dampen EMT in these cell types, we also observed some cell-line specific differences with respect to TGF-β1 activation of EMT characteristics. It is likely that the inherent genetic variations in the TGF-β1 signaling components of these cell types contribute to variations (33, 34). While PANC-1 has intact TGFBR2, it is mutated in MIA PaCa-2 (33). Consistently, a modest growth inhibitory effect of TGF-β1 was evident in PANC-1 but not MIA PaCa-2. Next, we noted exogenous TGF-β1 to activate the endogenous synthesis in PANC-1 but not MIA PaCa-2 (Fig. S12) and the auto-stimulatory effects in PANC-1 were attenuated by DZNep. Further, TGF-β1 phosphorylated Smad3 in PANC-1 but not significantly in MIA PaCa-2 (Fig. S12B). Moreover, the endogenous levels of TGF-β1 were significantly higher in PANC-1 than MIA PaCa-2 (Fig. S12B). Given the genetic aberrations in TGF-β1 signaling pathway in MIA PaCa-2, the mechanism by which TGF-β1 triggers mesenchymal characteristics in MIA PaCa-2 remains perplexing; however, it becomes increasingly clear that non-canonical TGF-β pathways, including the epigenetic pathways, could play prominent roles in deciding the EMT signature of PDAC cells. Further intriguing was that DZNep resisted TGF-β1-induced EMT characteristics at approximately equal magnitudes in both these cell types. These observations suggest that epigenetic reprogramming of miRNAs could likely play a dominant role in inhibiting TGF-β1 in PDAC cells. In addition to p-Smad2, DZNep suppressed p-Smad3 induced by TGF-β1 in PANC-1 and decreased endogenous p-Smad3 levels in MIA PaCa-2 and PANC-1 (Fig. S12B). DZNep also suppressed many of the TGF-β1-induced cellular EMT phenotypes, including their migratory and invasive behaviors in both cell types. Even in the presence of TGF-β1, DZNep consistently increased nucleoside analog cytotoxicity in PDAC cell lines regardless of whether TGF-β1 promoted further chemoresistance or not. Although the mechanism of how DZNep inhibits exogenous TGF-β1-induced EMT characteristics in pancreatic cancer cells warrants further investigation, overall these findings support the overriding potential of synthetic epigenetic reversal agents in inhibiting TGF-β-mediated EMT characteristics in PDAC.
The identification of profound reprogramming of miRNAs by DZNep uncovered several putative mechanisms involved in cytostatic growth control and EMT reversal in PDAC. For instance, DZNep induced the expressions of ~50 miRNAs by ≥1.5–10 folds. Of these miRs, several candidates have been reported earlier to confer anti-tumorigenic roles in various solid tumors (35–41). While they provide supportive evidence for DZNep’s growth control effects in PDAC, comparison of DZNep-induced miRNAs with downregulated miRNA datasets in PDAC further revealed which miRNAs are playing a role in the TGF-β1-induced EMT. Particularly, we focused on miR-663a and miR-4787-5p for their consistent downregulation in PDAC tissues and cell lines and upregulation by DZNep at various time points. Interestingly, miR-4787-5p is a novel miRNA with no studies available in literature. MiR-663a has been previously reported as a primate-specific miRNA silenced in other solid tumors by DNA methylation (42–44). This study provides primal evidence for silencing of miR-663/4787 in human PDAC due to histone methylation and for the use of DZNep as a pharmacological activator of miR-663/4787 expression for EMT reversal.
Intriguingly, the majority of DZNep-induced miRNAs, particularly miR-663a and miR-4787-5p (>90%), were high in GC content (Table S7). The mechanism of induction of GC-rich miRs by DZNep is unclear at present, but it is interesting to note that the 3′UTR region of the TGF-β1 mRNA, where most miRNAs bind and function, also has GC-rich regions (45). Further, miR-663a and miR-4787-5p reduced TGF-β1 protein levels without affecting TGF-β1 mRNA by directly binding to the TGF-β1 3′UTR and causing RNA interference. Even in presence of Actinomycin-D (transcription inhibitor), exogenous miR-663a or miR-4787-5p had no effect on TGF-β1 mRNA (data not shown) suggesting the lack of changes in TGF-β1 mRNA are not due to high turnover of the TGF-β1 mRNA. These findings are supportive of a physiological role for miR-663a and miR-4787-5p in regulating TGF-β1 function. Indeed, as this manuscript is completed, two independent groups showed miR-663 involvement in regulating TGF-β signaling in glioblastoma and thyroid carcinoma (46–48). Surprisingly, miR-663a and miR-4787-5p also attenuated TGF-β1-induced p-Smad2 which suggests their role in regulating other signaling components in addition to TGF-β1. Bioinformatics analyses failed to predict the involvement of any canonical TGF-β signaling players as targets for miR-663a or miR-4787-5p, and hence the mechanism of miR-663a and miR-4787-5p mediated suppression of p-Smad2 requires further investigation.
We suspect that several other DZNep-reprogrammed miRNAs are also capable of directly targeting the TGF-β1 3′UTR to RNA interference since DZNep induced a much stronger TGF-β1 RNA interference effect than either miR-663a or miR-4787-5p in PANC-1 cells. Recent findings in the laboratory showed DZNep to reduce the expression of TGF-β receptors and could provide a plausible explanation for DZNep’s antagonistic effects on exogenous TGF-β. While this is further explored in the laboratory, we also note that DZNep could target other key players (in addition to TGF-β) in the TGF-β signaling pathway (Table S8). Together, these scenarios provide support for why DZNep imparted stronger growth and metastasis inhibition effects on PANC-1 derived tumors in vivo than that observed with either miRNAs. Further, it is likely that the decreased metastatic load in DZNep/miR arms, at least in part, could be due to decreased size of primary tumors. However, we did not always find a correlation between tumor size and metastatic lesions (Fig. 7B–7G). For instance, miR-4787-5p tumors were relatively larger as compared with DZNep and miR-663a; however, it was as efficient, if not better, as them in suppressing metastasis. Nonetheless, at this point we do not have further evidence to explain the apparent discrepancies. Experimentally, we found no evidence for increased Caspase-3 cleavage in tumors from mice belonging to DZNep and miR groups (Fig. S13), whereas our in vitro data from this study (Fig. 6I & 6J) and our previous study on cultured pancreatic cancer cells showed DZNep to reduce cellular proliferation (25). These results indicate that the reduced tumor sizes noted with DZNep and miRs could be more likely attributed to reduced cell proliferation and not increased apoptosis. While the current study confirms the role of miR-663a and miR-4787-5p in growth control and EMT resistance in PDAC, the functions of numerous other PDAC-downregulated and DZNep-induced miRs remain to be interrogated.
The development of DZNep as a potential therapeutic compound may present some limitations. For example, DZNep exerted some nonspecific actions such as increasing the expression of certain potential oncomiRs (e.g., miR-10a and miR-10b) and decreasing the expression of certain potential tumor suppressor miRs (i.e., miR-181b). Further, DZNep’s anti-tumorigenic effects occurred only at lower micromolar concentrations, although this may not be a significant concern since most FDA-approved nucleoside analogs exhibit an effective therapeutic plasma concentration at this range. DZNep has also been reported to cause an unintended nephrotoxicity in rodents over long-term usage with some studies even suggesting DZNep can hinder normal developmental functions. While this could be very well attributed to DZNep’s global histone methylation altering effects, an attempt was made in this study to refine the applicability of histone methylation reversal agents by investigating its molecular targets and identifying candidates responsible for its beneficial actions. As the cause for cancer initiation and progression are multifactorial, it also becomes imperative to globally reprogram the cancer genome in a directed manner to desirably impart growth and EMT control. In this regard, both microRNAs and DZNep-like compounds offer a tremendous potential to future research in this direction. Identification of more favorable DZNep analogs exploiting desirably reprogrammed miRNAs or direct delivery of reprogrammed miRNAs into tumors offers two potential avenues for therapeutic development. The recent preclinical pharmacokinetic studies optimizing intravenous administration of DZNep for utilities in advanced-stage solid tumors are also very encouraging (49).
It is well known that EMT facilitates migration and invasion of tumor cells that leads to metastasis and development of chemoresistance. EMT is triggered by perturbations of the autocrine/paracrine TGF-β signaling towards late stage cancer. Hence, this pathway remains an attractive target of intervention to control EMT. Plasma TGF-β1 levels are also elevated in advanced pancreatic carcinoma patients who are presented with increased risk of poor prognosis (50). TGF-β1 levels are also significantly higher in patients not responding to chemotherapy as compared with the responding group (50). These clinical correlations suggest that TGF-β ligands and/or their receptors could be important targets in curtailing PDAC progression. Indeed, humanized monoclonal antibodies targeting TGF-β ligands and receptors, antisense oligonucleotides targeting TGF-β ligands, and small molecule inhibitors of TGF-β receptors have been developed and tested (5). While a few remain in phase I and II clinical trial evaluations, unfortunately, many of them have already been terminated due to low efficacies or high toxicities (5). So far, LY2157299, a small molecule inhibitor of TGFBR1, is the most advanced TGF-β signaling inhibitor currently under clinical development (5, 51). The approach used in this study provides a new direction for investigating small molecule inhibitors of TGF-β signaling in PDAC via microRNA reprogramming of cancer cells.
Supplementary Material
Implications.
The findings support the potential for synthetic histone methylation reversal agents to be included in future epigenetic-chemotherapeutic combination therapies for pancreatic cancer.
Acknowledgments
Financial Support: This work was supported by the award NIH 1R01CA188464 (RG)
Footnotes
Supplemental Materials and Methods & Supplemental Figures (Fig. S1–S13)
The authors disclose no potential conflicts of interest
References
- 1.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. The New England journal of medicine. 2014;371(11):1039–49. doi: 10.1056/NEJMra1404198. [DOI] [PubMed] [Google Scholar]
- 2.Satoh K, Hamada S, Shimosegawa T. Involvement of epithelial to mesenchymal transition in the development of pancreatic ductal adenocarcinoma. Journal of gastroenterology. 2015;50(2):140–6. doi: 10.1007/s00535-014-0997-0. [DOI] [PubMed] [Google Scholar]
- 3.Smith BN, Bhowmick NA. Role of EMT in Metastasis and Therapy Resistance. Journal of clinical medicine. 2016;5(2) doi: 10.3390/jcm5020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Javle MM, Gibbs JF, Iwata KK, Pak Y, Rutledge P, Yu J, et al. Epithelial-mesenchymal transition (EMT) and activated extracellular signal-regulated kinase (pErk) in surgically resected pancreatic cancer. Annals of surgical oncology. 2007;14(12):3527–33. doi: 10.1245/s10434-007-9540-3. [DOI] [PubMed] [Google Scholar]
- 5.Neuzillet C, de Gramont A, Tijeras-Raballand A, de Mestier L, Cros J, Faivre S, et al. Perspectives of TGF-beta inhibition in pancreatic and hepatocellular carcinomas. Oncotarget. 2014;5(1):78–94. doi: 10.18632/oncotarget.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Truty MJ, Urrutia R. Basics of TGF-beta and pancreatic cancer. Pancreatology : official journal of the International Association of Pancreatology. 2007;7(5–6):423–35. doi: 10.1159/000108959. [DOI] [PubMed] [Google Scholar]
- 7.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 8.Massague J. TGFbeta signalling in context. Nature reviews Molecular cell biology. 2012;13(10):616–30. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heldin CH, Vanlandewijck M, Moustakas A. Regulation of EMT by TGFbeta in cancer. FEBS letters. 2012;586(14):1959–70. doi: 10.1016/j.febslet.2012.02.037. [DOI] [PubMed] [Google Scholar]
- 10.Katsuno Y, Lamouille S, Derynck R. TGF-beta signaling and epithelial-mesenchymal transition in cancer progression. Current opinion in oncology. 2013;25(1):76–84. doi: 10.1097/CCO.0b013e32835b6371. [DOI] [PubMed] [Google Scholar]
- 11.de Caestecker MP, Piek E, Roberts AB. Role of transforming growth factor-beta signaling in cancer. Journal of the National Cancer Institute. 2000;92(17):1388–402. doi: 10.1093/jnci/92.17.1388. [DOI] [PubMed] [Google Scholar]
- 12.Kubiczkova L, Sedlarikova L, Hajek R, Sevcikova S. TGF-beta - an excellent servant but a bad master. Journal of translational medicine. 2012;10:183. doi: 10.1186/1479-5876-10-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowdhury S, Ammanamanchi S, Howell GM. Epigenetic Targeting of Transforming Growth Factor beta Receptor II and Implications for Cancer Therapy. Molecular and cellular pharmacology. 2009;1(1):57–70. doi: 10.4255/mcpharmacol.09.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo L, Zhang Y, Zhang L, Huang F, Li J, Wang S. MicroRNAs, TGF-beta signaling, and the inflammatory microenvironment in cancer. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015 doi: 10.1007/s13277-015-4374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiesslich T, Pichler M, Neureiter D. Epigenetic control of epithelial-mesenchymal-transition in human cancer. Molecular and clinical oncology. 2013;1(1):3–11. doi: 10.3892/mco.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danquah M, Singh S, Behrman SW, Mahato RI. Role of miRNA and cancer stem cells in chemoresistance and pancreatic cancer treatment. Expert opinion on drug delivery. 2012;9(12):1443–7. doi: 10.1517/17425247.2012.722079. [DOI] [PubMed] [Google Scholar]
- 17.McCleary-Wheeler AL, Lomberk GA, Weiss FU, Schneider G, Fabbri M, Poshusta TL, et al. Insights into the epigenetic mechanisms controlling pancreatic carcinogenesis. Cancer letters. 2013;328(2):212–21. doi: 10.1016/j.canlet.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu S, Ye D, Guo W, Yu W, He Y, Hu J, et al. G9a is essential for EMT-mediated metastasis and maintenance of cancer stem cell-like characters in head and neck squamous cell carcinoma. Oncotarget. 2015;6(9):6887–901. doi: 10.18632/oncotarget.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Deng L, Chen F, Yao Y, Wu B, Wei L, et al. Inhibition of histone H3K79 methylation selectively inhibits proliferation, self-renewal and metastatic potential of breast cancer. Oncotarget. 2014;5(21):10665–77. doi: 10.18632/oncotarget.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao RA, Dhele N, Cheemadan S, Ketkar A, Jayandharan GR, Palakodeti D, et al. Ezh2 mediated H3K27me3 activity facilitates somatic transition during human pluripotent reprogramming. Scientific reports. 2015;5:8229. doi: 10.1038/srep08229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tiwari N, Tiwari VK, Waldmeier L, Balwierz PJ, Arnold P, Pachkov M, et al. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer cell. 2013;23(6):768–83. doi: 10.1016/j.ccr.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 22.Oktyabri D, Tange S, Terashima M, Ishimura A, Suzuki T. EED regulates epithelial-mesenchymal transition of cancer cells induced by TGF-beta. Biochemical and biophysical research communications. 2014;453(1):124–30. doi: 10.1016/j.bbrc.2014.09.082. [DOI] [PubMed] [Google Scholar]
- 23.Tange S, Oktyabri D, Terashima M, Ishimura A, Suzuki T. JARID2 is involved in transforming growth factor-beta-induced epithelial-mesenchymal transition of lung and colon cancer cell lines. PloS one. 2014;9(12):e115684. doi: 10.1371/journal.pone.0115684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondo Y. Targeting histone methyltransferase EZH2 as cancer treatment. Journal of biochemistry. 2014;156(5):249–57. doi: 10.1093/jb/mvu054. [DOI] [PubMed] [Google Scholar]
- 25.Hung SW, Mody H, Marrache S, Bhutia YD, Davis F, Cho JH, et al. Pharmacological reversal of histone methylation presensitizes pancreatic cancer cells to nucleoside drugs: in vitro optimization and novel nanoparticle delivery studies. PloS one. 2013;8(8):e71196. doi: 10.1371/journal.pone.0071196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhutia YD, Hung SW, Krentz M, Patel D, Lovin D, Manoharan R, et al. Differential processing of let-7a precursors influences RRM2 expression and chemosensitivity in pancreatic cancer: role of LIN-28 and SET oncoprotein. PloS one. 2013;8(1):e53436. doi: 10.1371/journal.pone.0053436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim MP, Evans DB, Wang H, Abbruzzese JL, Fleming JB, Gallick GE. Generation of orthotopic and heterotopic human pancreatic cancer xenografts in immunodeficient mice. Nature protocols. 2009;4(11):1670–80. doi: 10.1038/nprot.2009.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinck AP, Archer SJ, Qian SW, Roberts AB, Sporn MB, Weatherbee JA, et al. Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2. Biochemistry. 1996;35(26):8517–34. doi: 10.1021/bi9604946. [DOI] [PubMed] [Google Scholar]
- 29.Subramanian G, Schwarz RE, Higgins L, McEnroe G, Chakravarty S, Dugar S, et al. Targeting endogenous transforming growth factor beta receptor signaling in SMAD4-deficient human pancreatic carcinoma cells inhibits their invasive phenotype1. Cancer research. 2004;64(15):5200–11. doi: 10.1158/0008-5472.CAN-04-0018. [DOI] [PubMed] [Google Scholar]
- 30.Rao ZY, Cai MY, Yang GF, He LR, Mai SJ, Hua WF, et al. EZH2 supports ovarian carcinoma cell invasion and/or metastasis via regulation of TGF-beta1 and is a predictor of outcome in ovarian carcinoma patients. Carcinogenesis. 2010;31(9):1576–83. doi: 10.1093/carcin/bgq150. [DOI] [PubMed] [Google Scholar]
- 31.Hasegawa S, Nagano H, Konno M, Eguchi H, Tomokuni A, Tomimaru Y, et al. A crucial epithelial to mesenchymal transition regulator, Sox4/Ezh2 axis is closely related to the clinical outcome in pancreatic cancer patients. International journal of oncology. 2016;48(1):145–52. doi: 10.3892/ijo.2015.3258. [DOI] [PubMed] [Google Scholar]
- 32.Liu L, Xu Z, Zhong L, Wang H, Jiang S, Long Q, et al. Enhancer of zeste homolog 2 (EZH2) promotes tumour cell migration and invasion via epigenetic repression of E-cadherin in renal cell carcinoma. BJU international. 2016;117(2):351–62. doi: 10.1111/bju.12702. [DOI] [PubMed] [Google Scholar]
- 33.Deer EL, Gonzalez-Hernandez J, Coursen JD, Shea JE, Ngatia J, Scaife CL, et al. Phenotype and genotype of pancreatic cancer cell lines. Pancreas. 2010;39(4):425–35. doi: 10.1097/MPA.0b013e3181c15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jazag A, Ijichi H, Kanai F, Imamura T, Guleng B, Ohta M, et al. Smad4 silencing in pancreatic cancer cell lines using stable RNA interference and gene expression profiles induced by transforming growth factor-beta. Oncogene. 2005;24(4):662–71. doi: 10.1038/sj.onc.1208102. [DOI] [PubMed] [Google Scholar]
- 35.Leung YK, Chan QK, Ng CF, Ma FM, Tse HM, To KF, et al. Hsa-miRNA-765 as a key mediator for inhibiting growth, migration and invasion in fulvestrant-treated prostate cancer. PloS one. 2014;9(5):e98037. doi: 10.1371/journal.pone.0098037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang L, Li X, Zhang X, Lv Z, He G, Zhao W, et al. MicroRNA-137, an HMGA1 target, suppresses colorectal cancer cell invasion and metastasis in mice by directly targeting FMNL2. Gastroenterology. 2013;144(3):624–35. e4. doi: 10.1053/j.gastro.2012.11.033. [DOI] [PubMed] [Google Scholar]
- 37.Ma F, Song H, Guo B, Zhang Y, Zheng Y, Lin C, et al. MiR-361-5p inhibits colorectal and gastric cancer growth and metastasis by targeting staphylococcal nuclease domain containing-1. Oncotarget. 2015;6(19):17404–16. doi: 10.18632/oncotarget.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun V, Zhou WB, Nosrati M, Majid S, Thummala S, de Semir D, et al. Antitumor activity of miR-1280 in melanoma by regulation of Src. Molecular therapy : the journal of the American Society of Gene Therapy. 2015;23(1):71–8. doi: 10.1038/mt.2014.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Q, Wei T, Shim K, Wright K, Xu K, Palka-Hamblin HL, et al. Atypical role of sprouty in colorectal cancer: sprouty repression inhibits epithelial-mesenchymal transition. Oncogene. 2015 doi: 10.1038/onc.2015.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, He X, Liu Y, Ye Y, Zhang H, He P, et al. microRNA-320a inhibits tumor invasion by targeting neuropilin 1 and is associated with liver metastasis in colorectal cancer. Oncology reports. 2012;27(3):685–94. doi: 10.3892/or.2011.1561. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Li T, Guo P, Kang J, Wei Q, Jia X, et al. MiR-424-5p reversed epithelial-mesenchymal transition of anchorage-independent HCC cells by directly targeting ICAT and suppressed HCC progression. Scientific reports. 2014;4:6248. doi: 10.1038/srep06248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y, Wang LL, Wang HX, Guo ZK, Gao XF, Cen J, et al. The epigenetically-regulated miR-663 targets H-ras in K-562 cells. The FEBS journal. 2013;280(20):5109–17. doi: 10.1111/febs.12485. [DOI] [PubMed] [Google Scholar]
- 43.Shi Y, Chen C, Zhang X, Liu Q, Xu JL, Zhang HR, et al. Primate-specific miR-663 functions as a tumor suppressor by targeting PIK3CD and predicts the prognosis of human glioblastoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20(7):1803–13. doi: 10.1158/1078-0432.CCR-13-2284. [DOI] [PubMed] [Google Scholar]
- 44.Zang W, Wang Y, Wang T, Du Y, Chen X, Li M, et al. miR-663 attenuates tumor growth and invasiveness by targeting eEF1A2 in pancreatic cancer. Molecular cancer. 2015;14:37. doi: 10.1186/s12943-015-0315-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Scotto L, Assoian RK. A GC-rich domain with bifunctional effects on mRNA and protein levels: implications for control of transforming growth factor beta 1 expression. Molecular and cellular biology. 1993;13(6):3588–97. doi: 10.1128/mcb.13.6.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu W, Xu S, Yao B, Hong M, Wu X, Pei H, et al. MiR-663 inhibits radiation-induced bystander effects by targeting TGFB1 in a feedback mode. RNA biology. 2014;11(9):1189–98. doi: 10.4161/rna.34345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Q, Cheng Q, Chen Z, Peng R, Chen R, Ma Z, et al. MicroRNA-663 inhibits the proliferation, migration and invasion of glioblastoma cells via targeting TGF-beta1. Oncology reports. 2016;35(2):1125–34. doi: 10.3892/or.2015.4432. [DOI] [PubMed] [Google Scholar]
- 48.Wang Z, Zhang H, Zhang P, Dong W, He L. MicroRNA-663 suppresses cell invasion and migration by targeting transforming growth factor beta 1 in papillary thyroid carcinoma. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015 doi: 10.1007/s13277-015-4653-y. [DOI] [PubMed] [Google Scholar]
- 49.Sun F, Lee L, Zhang Z, Wang X, Yu Q, Duan X, et al. Preclinical pharmacokinetic studies of 3-deazaneplanocin A, a potent epigenetic anticancer agent, and its human pharmacokinetic prediction using GastroPlus. European journal of pharmaceutical sciences : official journal of the European Federation for Pharmaceutical Sciences. 2015;77:290–302. doi: 10.1016/j.ejps.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 50.Vizio B, Novarino A, Giacobino A, Cristiano C, Prati A, Ciuffreda L, et al. Potential plasticity of T regulatory cells in pancreatic carcinoma in relation to disease progression and outcome. Experimental and therapeutic medicine. 2012;4(1):70–8. doi: 10.3892/etm.2012.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herbertz S, Sawyer JS, Stauber AJ, Gueorguieva I, Driscoll KE, Estrem ST, et al. Clinical development of galunisertib (LY2157299 monohydrate), a small molecule inhibitor of transforming growth factor-beta signaling pathway. Drug design, development and therapy. 2015;9:4479–99. doi: 10.2147/DDDT.S86621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.