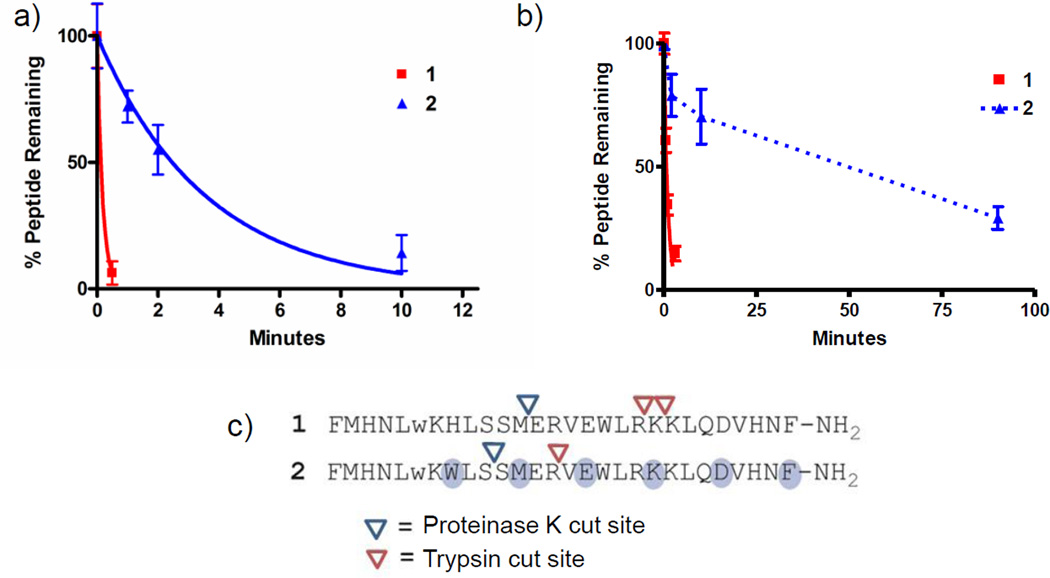
Figure 5. Evaluation of peptide stabilities in the presence of proteinase K or trypsin.
Peptides were incubated with (a) proteinase K or (b) trypsin using conditions described in the methods section. The amount of peptide remaining intact at indicated time points was measured using an HPLC-based assay. Data are presented as means (± SD) from a single experiment, with each condition assessed in duplicate. Results were fitted to an exponential decay model constrained to a plateau value of zero, which showed good agreement between the model and experimental data (r2>0.95), except for peptide 2 in the presence of trypsin, which was not evaluated using this model. Data points for peptide 2 in panel b are connected by a broken line to reflect this difference. The exponential decay model was used to provide estimated peptide half-lives for degradation experiments, except for peptide 2 in the presence of trypsin. Peptide half-lives are reported in Table 1. A replicate trypsin proteolysis experiment yielding similar data is presented in figure S4. (c) Amide bonds susceptible to protease-catalyzed hydrolysis were identified through characterization of peptide fragments appearing after incubation with protease using MALDI-TOF masss pectrometry.