Figure 3. The allosteric activation mechanism of SIRT1.
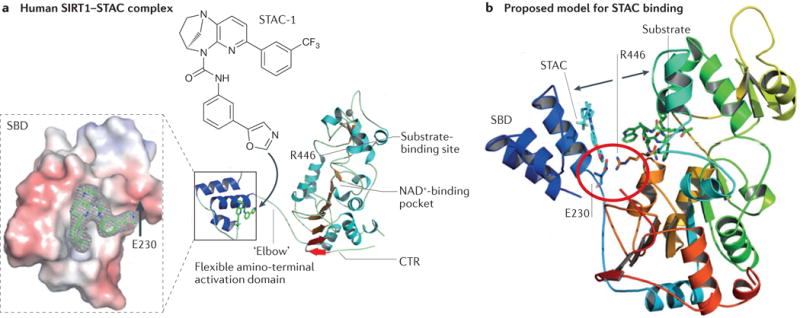
a| The structure of a truncated human sirtuin 1 (SIRT1) in complex with a synthetic sirtuin-activating compound (STAC) showing the binding site of STAC-1 at the amino-terminal STAC-binding domain (SBD). STAC binding promotes the interaction of the SBD with the central catalytic domain to lower the Km for the substrate, thereby increasing SIRT1 activity. The carboxy-terminal regulatory segment (CTR) stabilizes the deacetylase activity. Substitution of Glu230 (E230) with a Lys residue impairs enzymatic activation by reducing the electrostatic interaction between Glu230 on the SBD and Arg446 on the substrate-binding site. b | Model of how STACs activate SIRT1. Structural analysis and mutagenesis indicated that the amino acids Glu230 and Arg446 form a salt bridge (red circle) to facilitate binding of the amino terminus of SIRT1 with its catalytic core (movement indicated by arrows). The protein ribbon is rainbow-coloured from blue at the amino terminus to red at the carboxyl terminus. A STAC1 and an acetylated p53 peptide substrate are shown in cyan and green, respectively. Adapted from REF. 91, Nature Publishing Group.
