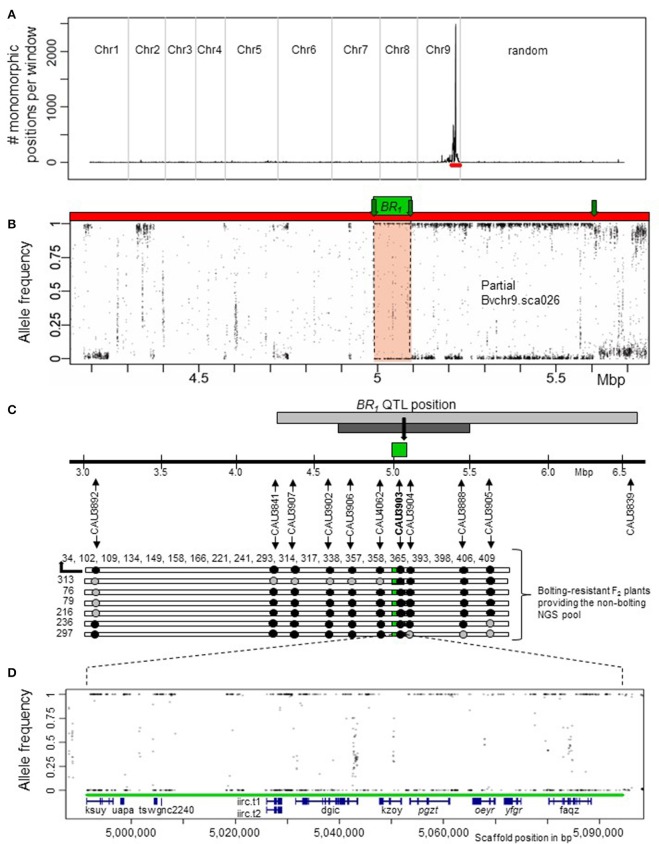
Figure 2.
Localization of the BR1 locus by a mapping-by-sequencing strategy. (A) A major peak was detected on chromosome 9 at scaffold Bvchr9.sca026 (red bar) by plotting the number of monomorphic positions of the br pool out of polymorphic positions of the b pool within a 200 kb sliding window and a step size of 100 kb. (B) Allele frequencies in the br pools based on polymorphic positions in the b pool at scaffold Bvchr9.sca026 (red). The genome-wide screening had revealed only a single, 103 kb region (green box) that was completely monomorphic in the br pool (allele frequency values 1 and 0 indicate monomorphic positions different and identical to RefBeet-1.1, respectively) at positions that were polymorphic in the b pool. This region is located at Bvchr9.sca026 between position 4,991,549 and 5,094,401. For the remaining genome positions, the allele frequencies differed from 1 and 0, whereby positions at unlinked regions showed on average allele frequencies of about 0.5 (not shown). Green arrows indicate recombination sites of bolting-resistant F2 plants. (C) Co-localization of the BR1 QTL and the physically mapped BR1 locus (green). Localization of the BR1 QTL (light gray), its confidence interval (dark gray) and QTL position (black arrow) is based on the sequence of the QTL flanking markers CAU3841 and CAU3839 (Pfeiffer et al., 2014) in relation to RefBeet-1.1. Genotypic data derived from 10 codominant CAU markers at the BR1 locus indicate crossover events around the BR1 locus in 6 out of 26 bolting-resistant F2 plants. The plant IDs of 20 plants homozygous for BR1: 34, 102, 109, 134, 149, 158, 166, 221, 241, 293, 314, 317, 338, 357, 358, 365, 393, 398, 406, 409. Black arrows show marker positions, black dots indicate marker positions that are homozygous for the allele derived from the bolting-resistant parent BETA 1773, gray dots indicate heterozygous positions. (D) Location of RefBeet-1.1 gene models (blue) within the physically mapped BR1 locus (green): ksuy.t1, uapa.t1, tswg.t1, nc2240, iirc.t1/t2, dgic.t1, kzoy.t1, pgzt.t1, oeyr.t1, yfgr.t1, faqz.t1. Details are given in Table 1. Genes in reverse orientation are written in italics.