Abstract
Long non-coding RNAs (lncRNAs) play important roles in several processes including control of gene expression. In Arabidopsis thaliana, a class of lncRNAs is produced by a specialized RNA Polymerase V (Pol V), which is involved in controlling genome activity by transcriptional gene silencing. lncRNAs produced by Pol V have been proposed to serve as scaffolds for binding of several silencing factors which further mediate the establishment of repressive chromatin modifications. We present methods for discovery and characterization of lncRNAs produced by Pol V. Chromatin Immunoprecipitation coupled with deep sequencing (ChIP-seq) allows discovery of genomic regions bound by proteins in a manner dependent on either Pol V or transcripts produced by Pol V. RNA Immunoprecipitation (RIP) allows testing lncRNA-protein interactions at identified loci. Finally, real-time RT-PCR allows detection of low abundance Pol V transcripts from total RNA. These methods may be more broadly applied to discovery and characterization of RNAs produced by distinct RNA Polymerases.
Keywords: non-coding RNA, ChIP, gene silencing, RNA Polymerase, RIP
1. Introduction
Long non-coding RNA (lncRNA), among other functions, is involved in directing chromatin modifications in order to control genes and transposons [1]. These modifications include de novo DNA methylation, histone modifications, and nucleosome positioning in a pathway known as RNA-mediated transcriptional gene silencing or RNA-dependent DNA methylation (RdDM) [2],[3],[4]. Mutations in components of the RdDM pathway can cause increased transposon activity, faulty DNA repair, or misregulation of genes [4],[5],[6].
In Arabidopsis thaliana, a class of lncRNAs involved in RdDM is produced by RNA Polymerase V, a specialized DNA dependent RNA polymerase [7]. Pol V transcripts are thought to create scaffolds on which several factors bind in order to control chromatin states [3],[8]. One of these factors is ARGONAUTE4 (AGO4), which binds to the Pol V C-terminal domain as well as to RNA and DNA [9],[10]. AGO4 is guided to chromatin by not only Pol V and its transcripts, but also by 24 nucleotide small interfering RNA (siRNA) [4],[8]. These siRNAs are generated from cleavage of double stranded transcripts produced by the coordinated activity of RNA Polymerase IV and RNA-dependent RNA Polymerase 2 [4],[11]. AGO4 associates with siRNA and is guided to chromatin based on the sequence specificity of the siRNA and the localization of Pol V [3],[5].
Another protein that binds to chromatin dependent on Pol V-produced lncRNA is SPT5-like (SPT5L) [12],[13],[14]. SPT5L works with AGO4 in a locus specific manner to direct the activity of the de novo DNA methyltransferase, DRM2 [2],[13].
Although DNA methylation is a major repressive chromatin modification established in RdDM, it is not the only one. Our recent work implicated Pol V-produced lncRNA in nucleosome positioning by indirectly recruiting a SWI/SNF chromatin remodeling complex via the IDN2 protein [15]. Being involved in the establishment of DNA methylation and repressive histone modifications as well as changes in nucleosome positioning, Pol V-produced lncRNA has broad effects on chromatin status and has been proposed to control genome activity [3]. Discovery of loci under control of Pol V-produced lncRNAs relies on finding genomic regions bound by Pol V [16],[17]. An additional approach is identifying binding sites of proteins recruited by Pol V transcripts [5]. Further investigation of the functional significance of those lncRNAs requires the ability to directly detect their presence and study their interactions with proteins [7],[9],[15],[16],[17]. We present methods, which allow the study of Pol V produced lncRNA as well as transcripts produced by other RNA polymerases. Although methods we show have only been tested with plant tissue, they should be applicable to the wide array of eukaryotic organisms.
2 Chromatin Immunoprecipitation (ChIP)
One of the main mechanisms used by lncRNA to control genome activity is by guiding proteins to specific genomic loci [3]. Detecting where these proteins are bound can give insights into the function of lncRNA [9],[13]. This approach may also be used in conjunction with high-throughput sequencing in order to discover new loci that are impacted by lncRNA [5],[16],[17]. Moreover, when performed in different mutant backgrounds, protein-DNA interaction assays may be used to further study molecular mechanisms involving these proteins.
Binding of specific proteins to DNA may be detected using Chromatin Immunoprecipitation (ChIP) [18]. In this method proteins and associated nucleic acids are precipitated using an antibody specific towards the protein of interest. DNA is further quantified and recovery of DNA above the background level is evidence of protein-DNA interaction.
An important step in ChIP is crosslinking with formaldehyde, which covalently fixes protein-DNA interactions but also makes it difficult to differentiate between direct and indirect interactions.
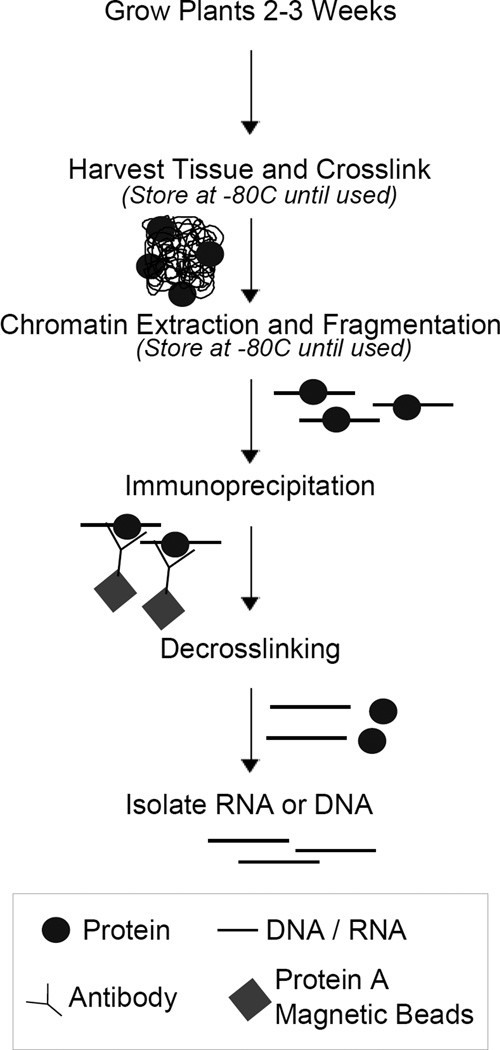
The specificity of particular antibodies may cause variability between experiments studying different proteins; it is advisable to optimize the amount of antibody used for each protein studied. Additionally, other key points, such as formaldehyde concentration or DNA fragmentation intensity, may be optimized depending on the protein studied. This method outlines basic procedures for crosslinking, chromatin isolation, immunoprecipitation, and DNA isolation (Figure 1), and has been successfully used for several proteins that bind chromatin in a way dependent on lncRNA [5],[7],[9],[13],[15],[16].
Figure 1. Overview of Chromatin-Immunoprecipitation (ChIP) and RNA-Immunoprecipitation (RIP).
DNA or RNA (black lines) is crosslinked to proteins (circles). Chromatin is extracted and fragmented (straight lines with circles). Fragments bound by the protein of interest are precipitated by an antibody (y-shaped) and magnetic beads (diamonds). RNA or DNA (straight lines) is purified from proteins (circles) and tested by real-time PCR.
2.1 Crosslinking
Successful ChIP usually requires a relatively large amount of starting material, thus 3g of approximately 2.5 weeks old Arabidopsis thaliana seedlings are used for each sample. Crosslinking is performed with formaldehyde (see Appendix A.1) [19]. In order to limit spontaneous decrosslinking, samples are kept on ice as much as possible. Samples are first washed once with ultrapure water to remove contaminants. Enough formaldehyde is added to completely cover the tissue and a vacuum is applied to infiltrate plants. To neutralize formaldehyde, glycine is added, mixed, and the plants are exposed to vacuum again. Samples are placed on ice and washed twice in ultrapure water. To make grinding easier, samples are lightly squeezed between paper towels in order to remove excess water and then frozen in liquid nitrogen. Samples may be stored at −80°C.
2.2 Chromatin Isolation
Frozen crosslinked plant material can now be directly used to isolate chromatin. Each step should be performed on ice. Samples are ground in liquid nitrogen to a fine powder and resuspended gently in ice cold Honda buffer (supplemented with DTT, PMSF, and Plant Protease Inhibitors - see Appendix B.3). The resuspended powder is filtered through two layers of Miracloth, and the flow-through is collected in an empty tube. To increase yield, the used Miracloth is washed in Honda buffer and the resulting solution is filtered through a clean Miracloth and the two filtrates combined. Nuclei are collected by centrifugation and washed several times with cold Honda buffer to remove cellular debris. After washes, the nuclei are resuspended in Nuclei Lysis Buffer (see Appendix A.1 and B.3).
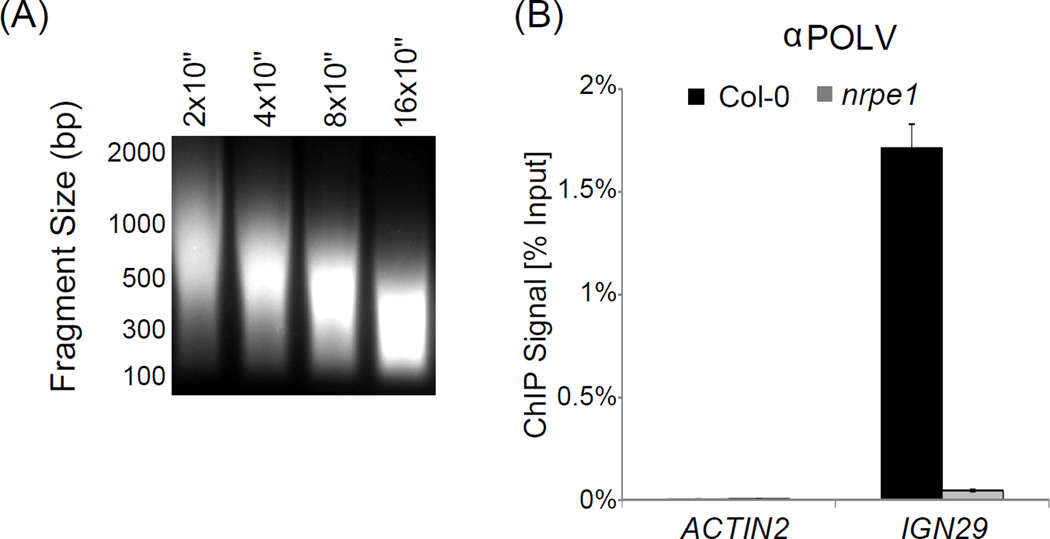
Nuclei are lysed and chromatin is fragmented by sonication on ice to an approximate average DNA length of between 250 and 500 bp. The fragmentation intensity should be experimentally optimized for a specific sonicator instrument prior to performing ChIP experiments (See Fig. 2A). Sonication optimization may be performed by skipping immunoprecipitation (steps 23–33) in our ChIP protocol (Appendix A.1) and by checking the DNA fragment sizes by gel electrophoresis. Average fragment length of sonicated DNA is an important variable since it limits resolution of the entire ChIP assay.
Figure 2. ChIP.
(A) DNA fragmentation after sonication. Fragmentation can be checked by gel electrophoresis after decrosslinking. Optimal fragmentation can be determined by varying the sonication time as shown.
(B) ChIP-based detection of DNA bound by NRPE1, the largest subunit of Pol V. DNA binding is seen as enrichment in Col-0 wild-type compared to nrpe1 mutant (which serves as a background level control) at the IGN29 locus. ACTIN2 serves as an unbound loading control to check specificity of immunoprecipitation. Bars represent average real time PCR signal normalized to inputs in Col-0 wild-type (black) and nrpe1 (gray). Error bars represent standard deviations from three PCR amplifications.
After fragmentation, nuclear debris is pelleted by centrifugation and the supernatant containing fragmented chromatin is kept. The large amount of starting material means that aliquots can be taken and kept at −80°C after flash freezing in liquid nitrogen, providing several ChIP experiments from one chromatin isolation step. A smaller aliquot from each sample should also be made and stored for input controls.
2.3 Immunoprecipitation and Elution of DNA
Immunoprecipitation is done with magnetic beads conjugated to protein A (Dynabeads Protein A). In preparation for immunoprecipitation, the magnetic beads are washed and resuspended in B/W buffer supplemented with PMSF (see Buffers Appendix B.3). All steps are performed on ice or at 4°C. Chromatin aliquots are thawed on ice and diluted with ChIP Dilution Buffer to reduce the concentration of SDS (see Appendix A.1 and B.3). The prepared magnetic beads are added, as is the antibody directed against the protein of interest. Immunoprecipitation is performed at 4°C rotating overnight. In addition to samples with antibody, samples without antibody can be included to determine the nonspecific background pulled-down directly by the beads.
The next day, the inputs are thawed on ice. Meanwhile, the beads are washed in B/W Buffer to remove non-precipitated chromatin. Another wash with TE is performed during which the beads are transferred to new tubes in order to reduce background by DNA bound nonspecifically to the tubes. After the washes, crosslinked protein-DNA complexes are eluted in Elution Buffer (see Buffers Appendix B.3) at 65°C. Inputs are also prepared by adding a small amount of chromatin to Elution Buffer. After transferring the eluate to a new tube, a second elution step is done to increase the yield of recovered DNA, and the eluates are combined. Inputs and ChIP samples are digested with Proteinase K at 60°C overnight. Extended heat treatment also reverses formaldehyde-induced crosslinks, which allows subsequent purification of DNA.
2.4 DNA Isolation
DNA is purified by phenol extraction (see Appendix A.1). After centrifugation the aqueous phase (top layer) is transferred to a new 1.5ml tube. Subsequent extraction with chloroform-isoamyl alcohol helps to remove traces of phenol in the sample, which is essential if samples are used for library preparation. DNA is precipitated with 96% Ethanol in the presence of GlycoBlue (or a different carrier compatible with downstream applications) and washed with 70% Ethanol. The supernatant is carefully removed and, after air drying, the pellet is resuspended in water. Alternately, if samples are to be used solely for real-time PCR analysis, 10% Chelex may be added directly to the magnetic beads after the final wash with TE [20]. Chelex should be added using a pipette tip cut at the 100µl mark while mixing the Chelex in between the addition to each sample. The tip is cut to ensure equal amounts of beads are added to each sample. Samples can then be incubated at 99°C for ten minutes. Proteinase K is then added and samples are incubated at 65°C for 2 hours followed by 95°C for ten minutes. Samples should be centrifuged at maximum speed for 1 minute and the supernatant taken to avoid carryover of Chelex before PCR analysis.
2.5 Analysis and Interpretation
Purified immunoprecipitated DNA may be analyzed by real-time PCR (section 4.2). Signal levels are usually calculated relative to input for every specific locus using a relative delta-cT method [21],[22]. Several controls are required to conclusively demonstrate protein binding to DNA.
Background level control shows signal that does not originate from specific antibody-epitope interactions. If a protein-specific antibody is used, a knock-out mutant line entirely lacking the protein of interest should be used as a background level control (Figure 2A – nrpe1). Alternatively, when an epitope tag-specific antibody is used to detect a tagged protein, a line which does not express the epitope-tagged protein also provides reliable background level information.
Loading control shows if technical issues arise during sample preparation. This has to be a locus where signal levels should be identical between samples. It can be beneficial to use more than one loading control locus. This control can either be loci where the protein does not bind (Figure 2B – ACTIN2) or loci where the protein is known to bind in all samples. Signal levels from the loading control may sometimes be used for further normalization of ChIP results.
No antibody control, in which ChIP is performed without an antibody or by using non-specific antibodies. It shows signal that does not originate from antibody interactions but is non-specifically carried over on the beads. Because most antibodies show non-specific interactions, this control often underestimates background levels and should not be used as a replacement to a background level control described above.
Positive control, which is a locus tested in all biological samples and based on pre-existing information is known to be bound by the protein of interest. This control is necessary if samples will be tested using high-throughput sequencing.
Protein binding to DNA is conclusively demonstrated if specific signal is detected above the background level signal and loading controls show no significant differences between biological samples (Figure 2B). Additionally, specific signal should be significantly higher than no antibody controls and a positive control should also demonstrate signal above the background level. Every ChIP experiment should be performed in at least three independent biological repeats (see Fig. 5A in [15] for an example).
2.6 High-throughput sequencing
Purified immunoprecipitated DNA, which has been successfully tested on specific loci using real-time PCR, may be used to discover new loci bound by the protein of interest by high-throughput sequencing. Samples used for high-throughput sequencing should have high signal to background proportion on known binding sites, ideally higher than 8×. If this proportion is lower, data analysis may be difficult and therefore the ChIP protocol should be optimized to increase signal to background ratio. Sequencing libraries can be generated using the Illumina ChIP-seq Library Preparation protocol. Detailed description of library generation is beyond the scope of this paper and usually is performed by specialized facilities. Analysis of ChIP-seq is discussed in section 5.
3. RNA-Immunoprecipitation (RIP) of Pol V Transcripts
Using a similar strategy as ChIP, protein-RNA interactions can be detected. When applied to studying interactions between proteins and Pol V-produced non-coding transcripts they provide a link between a protein’s effect on chromatin and the ability to bind lncRNA. As for ChIP, binding is manifested as enrichment between conditions/genotypes when analyzed by real-time RT-PCR. Protein-RNA interactions are covalently fixed using reversible chemical crosslinking. RIP follows the same steps as ChIP: crosslinking, chromatin preparation and fragmentation, immunoprecipitation, and nucleic acid isolation (Figure 1). Differences include enzymatic elimination of DNA and precautions against loss of RNA.
This approach shares all the limitations of ChIP, including limited resolution and inability to conclusively distinguish direct from indirect interactions. It is however much more straightforward than CLIP [23] and may successfully be applied for specific questions, especially if proper controls are available. The protocol described below and in Appendix A1 has been applied to study interactions of Pol V-produced lncRNAs with several proteins [9],[12],[15].
3.1 Procedure
Crosslinking is performed exactly as described for ChIP and crosslinked tissue can be used interchangeably between these methods. Chromatin isolation is also performed as described for ChIP with the addition of an RNase inhibitor to most buffers to prevent RNA degradation (see Buffers Appendix B.3). Immunoprecipitation is also performed as in ChIP, with a few minor modifications (see Appendix A1). Inputs are prepared by adding undiluted chromatin to Elution Buffer, and the following steps are performed with both inputs and precipitated samples. Two elution steps are performed to retrieve RNA, the first is performed at room temperature for ten minutes, while the second is performed at 65°C for ten minutes. Proteinase K digestion is done at 55°C for one hour 15 minutes. RNA is isolated by extraction with acidic phenol:chloroform:isoamyl alcohol (pH 4.3) followed by centrifugation and ethanol precipitation as in ChIP. After final wash in 70% ethanol it is critical to remove as much of the supernatant as possible and air dry the pellet for as short as possible (preferably 2 min.).
3.2 Analysis and Interpretation
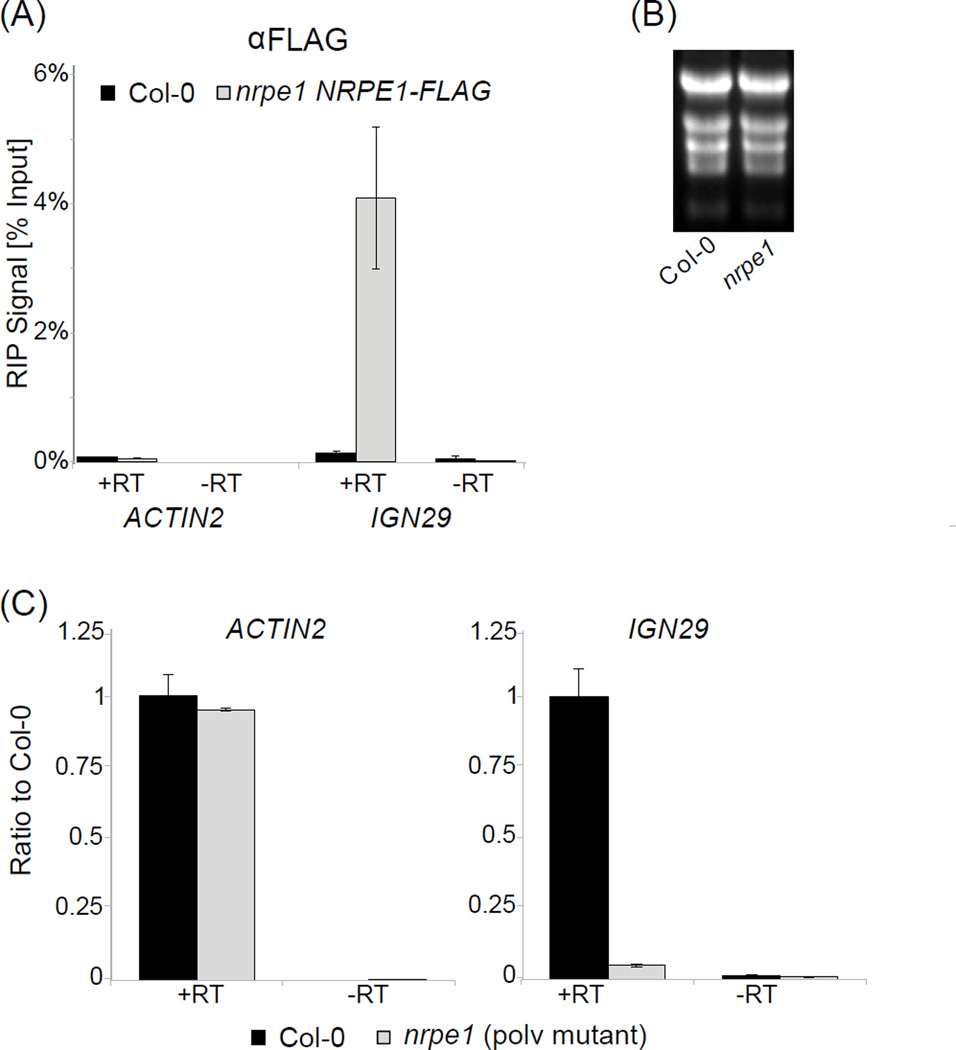
Using the reverse transcription method described in section 4, precipitated RNA should immediately be converted to cDNA (after DNase I digestion) and then analyzed by real-time RT-PCR. Evidence of binding to RNA can be established based on controls as described for ChIP (Section 2.5). If applied to studying a specific class of RNA, it is important to include an additional control, which allows determining background interaction of non-specific RNAs. In the case of Pol V-produced lncRNA this control may be a knock-out mutant in a Pol V subunit, which lacks Pol V transcripts and any detected signal may be attributed to RNAs produced by other RNA polymerases. A critical control is RT-PCR without reverse transcriptase (no RT) to detect DNA contaminations, which are the most common technical problem in RIP. An additional consideration is that in contrast to ChIP, signal levels in RIP may vary dramatically between tested loci due to differences in transcription levels. It is therefore possible that background signal levels detected at an unrelated locus may be much higher than specific signal. Input levels can provide additional information on RNA levels, however RNA obtained from input samples is often difficult to amplify. Normalization to inputs may be applied during analysis as shown in Figure 3A. Due to potential quality issues and variations in total RNA levels between samples, inputs should be examined before deciding to normalize. For example, Pol V produced lncRNAs are eliminated in the Pol V mutant (nrpe1) making normalization to inputs in nrpe1 impossible. For comparing independent biological replicates it is often necessary to normalize to wild type controls (see Fig. 1A in [15] for an example).
Figure 3. RIP and total RNA RT-PCR.
(A) RIP-based detection of RNA bound by Pol V. RNA binding is seen by enrichment between FLAG tagged Pol V subunit (nrpe1 NRPE1-FLAG) compared to Col-0 wild-type (which serves as a background control) at IGN29 [15]. ACTIN2 serves as an unbound loading control to check efficiency of immunoprecipitation. Samples without reverse transcriptase (-RT) are used to check for signal resulting from DNA contamination. Bars represent average real time RT-PCR signal normalized to inputs. Error bars represent standard deviations from three PCR amplifications.
(B) Quality of RNA used in RT-PCR. RNA quality may be seen by gel electrophoresis as shown.
(C) Non-coding transcripts are seen by enrichment between Col-0 wild-type and the Pol V mutant (nrpe1) at IGN29 [15]. ACTIN2 serves as a loading control. Samples without reverse transcriptase (-RT) are used to check for signal resulting from DNA contamination. Bars represent average RNA signal as a ratio of mutant (nrpe1 – gray) to Col-0 wild-type (black). Error bars represent standard deviations from three PCR amplifications.
4. Real-Time RT-PCR of low abundance lncRNA
Following RIP, RNA is converted to cDNA and amplified by real-time PCR. In addition to analyzing RIP, this method may be used for detection of lncRNAs in total RNA samples. Long non-coding RNAs, including Pol V transcripts, are generally transcribed at low levels (some approximately 10,000 fold lower than ACTIN2), causing them to be difficult to detect. This method is sensitive enough to reliably and quantitatively detect Pol V transcripts and other low abundance RNAs. The protocol involves a DNase digestion to eliminate DNA, reverse transcription and real-time PCR. We recommend using random oligonucleotides as primers, however if strand-specificity is required, locus-specific primers should be used. The method we describe has been used to detect and quantify several Pol V-produced lncRNAs [5],[15].
4.1 RNA Isolation and cDNA Synthesis
For detection of Pol V transcripts from total RNA, RNA isolation is performed with the RNeasy Plant Mini Kit from Qiagen. The optional on-column DNase I digestion step is followed to eliminate contaminating DNA. Alternately, RIP samples from section 3 are used. Quality of total RNA should be tested using denaturing agarose electrophoresis (Fig. 3B) or using a Bioanalyzer. Quality of immunoprecipitated RNA is very difficult to assay directly at this step, since expected amounts of RNA recovered from RIP are below detection thresholds of currently available methods.
One microgram of total RNA is sufficient for each reverse-transcription reaction to reliably detect Pol V transcripts. To further eliminate any DNA contamination, RNA is treated with Turbo DNase in the presence of Ribolock RNase Inhibitor. RNA is then separated into two reactions per genotype: half of the RNA is transferred to a new tube to be used as a no-reverse-transcriptase control (no RT), while the other half of the sample is transferred to a separate tube for the RT reaction. To denature and anneal primers, the RNA is mixed with random primers and dNTPs and incubated at 65°C followed by at least one minute on ice. For reverse-transcription, DTT, 5 × First Strand buffer, Ribolock, and Superscript III Reverse-Transcriptase are added. In case of the no-RT controls, water is added in place of Superscript III. Reverse transcription is performed at 50°C.
A critical technical issue is avoiding DNA contaminations, especially originating from previously handled PCR products. We recommend using filtered tips, disposable gloves and working in clean workspace to avoid possible contaminations.
4.2 Real-Time PCR of Long Non-Coding RNA
For real-time PCR, a small volume of the cDNA is added to a PCR reaction mix, which in addition to a Hot Start Taq Polymerase, corresponding buffer, magnesium, and dNTPs, also contains SYBR Green – a dye specific towards double-stranded DNA which allows real-time quantification of the PCR product. Depending on the real-time PCR instrument being used, it may be also necessary to add an internal reference dye. The reaction is run with one step at 95°C, followed by 40 cycles of 95°C, 55°C, and 72°C. It is recommended to finish each reaction with a melting curve to determine primer specificity and detect possible contaminations contributing to the signal.
4.3 Analysis and Interpretation
Obtained results should be analyzed using relative delta-cT method between samples [21],[22], which is sufficient for comparing signal levels generated with a primer pair in various biological samples but does not allow quantitative comparison between different primer pairs. If different primer pairs have to be compared, data should be analyzed using a standard curve.
If detecting Pol V transcripts in total RNA, enrichment between wild-type and a mutant in a Pol V subunit (nrpe1) indicates presence of lncRNA (Figure 3C). This assay may be used to show lncRNA production or quantitatively compare lncRNA levels between genotypes. If RIP samples are used, protein-RNA interactions can be seen as discussed in section 3.2.
For proper data interpretation, the possibility of DNA contaminations should be excluded by performing controls without reverse transcriptase. We recommend performing every experiment in three biological repeats and testing every biological repeat with three PCR amplifications. In ChIP and RIP experiments it is common that overall signal levels are variable between independent experiments. If it is the case, every repeat should be normalized to wild type prior to calculating averages and standard deviations for all available biological repeats (see [15] for an examples).
5. ChIP-Sequence Analysis
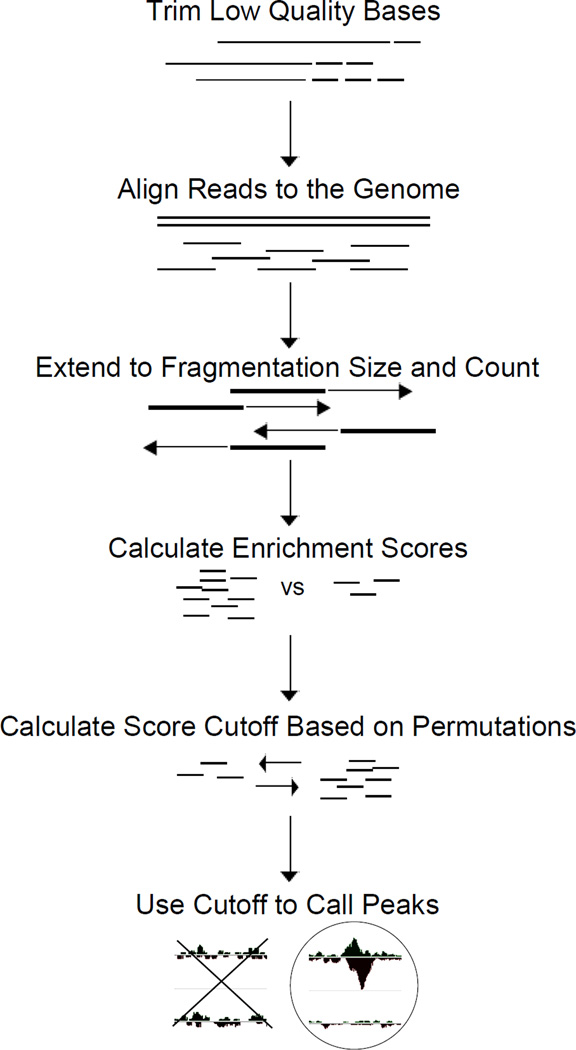
ChIP-seq is performed to identify protein binding sites throughout the genome. In order to generate a list of binding locations, the raw sequencing reads must first be processed in a way that accurately maps to the Arabidopsis genome while controlling for quality. After reads have been mapped, enrichment scores can be generated and utilized to obtain genomic coordinates of binding sites (Figure 4). Several different algorithms exist for each step and decisions on the use of each algorithm can be difficult; each has advantages and individual decisions must be made specific to the type of protein studied and the question being asked. Presented here (see Appendix A.3) is a simple data analysis pipeline based on published work [5], presented in a way which should provide a starting point to biologists and allow further refinement for a specific dataset. We use the peak-calling algorithm, CSAR [24], because it was built specifically around the Arabidopsis genome and from our experience has a high rate of discovery with a low number of false positives. Several aligners can be used and our experiences with each have been excellent. For simplicity sake we use SOAP2 [25] because the output format can directly be read into CSAR.
Figure 4. ChIP-Sequencing Analysis.
Analysis of sequencing reads from ChIP involves quality control, mapping, and peak calling. Low quality reads and bases (dashed lines) are removed from analysis to improve alignment. Remaining reads are aligned to the genome. Reads are extended to the fragmentation size obtained in sonication to more precisely map binding sites (arrows). Enrichment scores and permutations are used to obtain statistical significance for enriched genomic regions.
5.1 Read Trimming and Alignment
Although sequencing generally produces quality reads, it may be useful to trim any low quality bases from either end. There are several ways to do this, but ConDeTri [26] provides a simple method of trimming N’s from the 5’ end and low quality bases from the 3’ end of reads.
In order to align reads to the genome efficiently, alignment algorithms generally require an index of the reference genome sequence. Alignment packages usually include a separate algorithm for index creation. The Arabidopsis genome sequence can be downloaded from the TAIR website (ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR10_genome_release/TAIR10_chromosome_files/TAIR10_chr_all.fas) and indexed using 2bwt-builder in the SOAP2 package [25]. Index files will be given the suffix “.index”. Alignments can now be performed using the SOAP2 aligner.
5.2 Peak Detection
Once mapping is complete, binding sites are called based on regional read enrichment in wild-type compared to mutant. Although a knockout mutant represents the true background signal obtained during the ChIP experiment, inputs may also be used to control for sequence or amplification bias during library generation. Peak calling through enrichment scores can be done using CSAR which was built and tested around the Arabidopsis genome. First the mapped reads are loaded in R in a format that CSAR is able to recognize. Reads are then artificially extended to the fragmentation length obtained from sonication, and the number of reads at each base is counted for both strands. To remove PCR amplification errors, only unique reads are kept. Scores are calculated by comparing two samples (i.e. wild-type versus mutant) and regions with enrichment are kept. Significant binding sites are determined by establishing an arbitrary cutoff score at a specified false discovery rate (FDR). Generally an FDR of 0.001 provides high quality peaks, but peak lists at various FDR values can be generated. The final list will have chromosomal coordinates, the enrichment scores, and the length of peaks included in the file.
6. Conclusions
The activity of lncRNA can be studied through a combination of techniques focused on indentifying loci controlled by RdDM. New targets of transcriptional gene silencing can be detected by ChIP-seq for proteins that bind chromatin in a way dependent on lncRNA. These loci can then be directly tested for lncRNA production and protein-RNA interactions.
Additionally, by using a combination of these methods in various mutant backgrounds and with antibodies for different proteins, molecular mechanisms of RNA-mediated silencing pathways can be deciphered. As knowledge of lncRNA expands, methods such as these will become increasingly important to study its role in the regulation of genome activity.
Acknowledgments
We thank Brian Gregory and Qi Zheng for sharing expertise in high throughput sequencing data analysis. This work has been supported by National Science Foundation grant MCB 1120271 to A.T.W. and Austrian Science Fund (FWF) fellowship J3199-B09 to G.B.
Appendix A: Step-by-Step protocols
A.1 ChIP and RIP
Note: Use filter tips when working with RNA.
Note: Keep samples on ice as much as possible.
Harvest 3g of above ground tissues from 2.5 week old seedlings and place in 50ml tubes on ice with a hole punched in lids with scissors.
Rinse once with H2O.
Add 0.5% formaldehyde so that tissue is immersed (up to 35ml).
- Apply vacuum at 85 kPa (~25in. Hg) below standard atmospheric pressure for 2 minutes. Release and reapply for 8 minutes.Note: Ensure a hole is placed in each lid or tubes may break from the pressure.
Add 1.25ml 2M glycine. Mix and apply vacuum at 85 kPa (~25in.Hg) for 1 minute. Release and reapply for 4 minutes.
Rinse twice in H2O, squeeze dry between paper towels, wrap in aluminum foil, and freeze in liquid nitrogen.
- Store at −80°C.Note: Crosslinked tissue can be used for either ChIP or RIP.
- Prepare 30ml Honda Buffer per sample. For every 30ml Honda Buffer, add 150µl 1M DTT, 300µl 100mM PMSF, and 300µl Plant Protease Inhibitor. For RIP, also add 240u Ribolock RNase Inhibitor.Note: For all RNA work solutions should be prepared with RNase-free water. We recommend using fresh miliQ water without any additional treatments.
Place 15ml and 10ml prepared Honda Buffer in 50ml tubes on ice. Keep leftover buffer on ice as well.
- Grind frozen crosslinked tissue in liquid nitrogen into a fine powder and resuspend gently in 15ml prepared Honda Buffer. Keep on ice.Note: Ensure that all large chunks are dissolved and that a fine powder is present in the buffer.
Cut two 6cm wide strips of Miracloth and form one into a funnel with two layers between outside and inside of the funnel.
Filter the resuspended tissue through funnel into empty 50ml tube.
- Invert filter and place in prepared 10ml Honda Buffer. Resuspend as much tissue as possible by swirling Miracloth in buffer.Note: Washing the Miracloth increases chromatin yield
Using the second strip of Miracloth from step 11, form another funnel like the first and filter washed sample from step 13, combining the flow through with the previous one.
Centrifuge at 3000 g for 7.5 minutes at 4°C and discard supernatant.
Resuspend pellet with 1ml prepared Honda Buffer and transfer to a 1.5ml tube.
Centrifuge at 1900 g for 5 minutes at 4°C. Discard supernatant.
- Wash twice with 1ml prepared Honda Buffer by centrifugation at 1900 g for 5 minutes at 4°C. If the pellet is still green perform one additional washing step.Note: Use leftover Honda Buffer from step 9 for steps 16 and 18.
Resuspend nuclei gently in 550µl freshly prepared cold NLB (with 88U Ribolock for RIP).
- Sonicate on ice 8 times for 10 seconds with 1 minute pauses at power setting 1. (Fisher Scientific Dismembrator - Adapt number of times based on the instrument used until chromatin is fragmented to approximately 250bp to 500bp on average).Note: It is important to keep samples on ice during sonication to avoid overheating and possible protein degradation or loss of crosslinks.
Centrifuge samples in a microcentrifuge at maximum speed for 10 minutes at 4°C. Transfer supernatant to a new 1.5ml tube.
- Take 100µl aliquots and flash freeze in liquid nitrogen. Also freeze one 10µl aliquot for inputs. Store at −80°C.Note. While the preceding steps are specific for plant material, the remainder of the protocol may be applicable to chromatin samples obtained from other organisms.
Prepare 5ml B/W buffer by adding 50µl PMSF (and 200U Ribolock for RIP).
Using magnetic rack, wash 40µl per sample Dynabeads Protein A three times with 1ml B/W buffer from step 23. Resuspend in 110µl B/W from step 23 per sample.
Combine one 100µl aliquot of chromatin, 900µl ChIP Dilution Buffer (with Ribolock for RIP - see Appendix B.3), 100µl washed magnetic beads, and 2.5µg – 5µg antibody.
- Rotate at 4°C over-night.Note: Amount of antibody and incubation time can be adjusted depending on the protein studied or the antibody used. In our experience, 2.5µg – 5µg antibody works well for most applications. Magnetic beads may be replaced with Protein A Agarose beads blocked with salmon sperm DNA, which give lower background levels for some antibodies; however, due to the presence of blocking DNA, samples obtained using those beads cannot be used for sequencing
Prepare 5ml per sample B/W buffer by adding 50µl PMSF (and 200U Ribolock for RIP).
-
For ChIP wash the samples three times with 1ml B/W with rotation at 4°C for 5 minutes between washes. Follow with one wash in TE.
For RIP wash twice with 1ml B/W by inverting, magnetic separation, and removal of supernatant, followed by twice with 1ml B/W and rotating at 4°C for 5 minutes between washes.Note: Use a magnetic separator buried in ice during washes to keep samples cold. After the last wash transfer samples to new 1.5ml tubes, remove supernatant, and add 55µl Elution Buffer (with 22U Ribolock for RIP).
Prepare 1% input samples by adding 1µl chromatin extract to 110µl Elution Buffer (with 44U Ribolock for RIP).
- Vortex all samples briefly and incubate them at 65°C for 20 minutes for ChIP (room temperature for 10 minutes for RIP).Note: Vortexing vigorously helps elute the sample from the beads.
- Place on magnetic separator, transfer supernatant to a new 1.5ml tube and add 55µl Elution Buffer to beads (with 22U Ribolock for RIP).Note: Input samples do not need to be transferred
Incubate at 65°C for 20 minutes (10 minutes for RIP), vortex, place on magnetic separator, and transfer supernatant to previous eluate.
Add 20µg RNA-grade Proteinase K and incubate at 60°C overnight for ChIP (55°C for 1 hour 15 minutes for RIP).
Add 110µl room temperature phenol:chloroform:isoamyl-alcohol 25:24:1 (pH 7.5 for ChIP, pH 4.3 for RIP).
Vortex well and centrifuge at maximum speed for 5 minutes.
Transfer 100µl aqueous phase to new 1.5ml tube.
For ChIP repeat steps 35–37 with 24:1 chloroform:isoamyl-alcohol.
Add 60µg GlycoBlue (or an equivalent amount of a different carrier compatible with downstream applications), 10µl 3M sodium acetate pH 5.3, and 250µl 96% ethanol.
Incubate at −80°C for 2 hours (overnight for RIP).
Centrifuge at maximum speed for at least 30 minutes at 4°C. Discard supernatant.
Add 300µl 70% ethanol and centrifuge at maximum speed for 10 minutes at 4°C. Discard supernatant. If traces of supernatant are left on tube walls, centrifuge again briefly and remove leftovers of the supernatant.
Air dry pellet for 2 minutes and resuspend in 30µl H2O for ChIP (12µl RNase-free H2O for RIP). ChIP samples may be stored for up to a year at −20°C, RIP samples may be stored at −80°C for a couple of days, however due to low stability of RNA should be processed as soon as possible.
- ChIP DNA can be checked for enrichment of DNA levels between samples by real-time PCR or used for library generation and sequencing. RIP RNA can be reverse transcribed and used in quantitative PCR as in A.2.Note: Alternately, if ChIP DNA will not be used for sequencing, steps 29–43 can be replaced with the following protocol using Chelex. Cut a pipette tip at 100µl and add 100µl 10% Chelex-100 to the beads. Prepare 1% inputs by adding 1µl of chromatin to 100µl 10% Chelex-100. Vortex well and incubate at 99°C for ten minutes. Cool samples to 65°C and add 20µg Proteinase K, incubating at 65°C for two hours. Deactivate Proteinase K at 95°C for ten minutes and spin samples at max speed for 1 minute. Supernatant can be diluted and analyzed by real- time PCR as in step 44.
A.2 Real-Time RT-PCR of low abundance lncRNA
Note: Use filtered tips, disposable gloves and clean workspace when working with RNA.
- Isolate RNA using the Qiagen RNeasy Plant Mini Kit following instructions with the optional on-column DNase digestion. RIP samples from A.1 can also be used.Note: Total RNA may be stored at −80°C up to 2 weeks.
To 1–5µg total RNA or the entire RIP sample from A.1 step 43, add 3units Turbo DNase, 24 units Ribolock, and H2O up to 12µl.
Incubate at 25°C 30 minutes.
Add 3µl 25mM EDTA.
Incubate at 65°C 10 minutes.
Transfer 1/2 RNA to a new tube for no-RT control and transfer 1/2 RNA to a new tube for the RT reaction.
Add 0.4µl 500ng/µl random primers (Invitrogen) and 1µl 10mM dNTPs.
Incubate at 65°C 5 minutes. Keep on ice for at least 1 minute.
Add 1µl 0.1M DTT, 4µl 5x First Strand Buffer, 1µl 40U/µl Ribolock, and 1µl Superscript III Reverse Transcriptase (substitute water for Superscript III in no-RT controls). Add H2O to a total volume of 20µl.
- Incubate 25°C for 5 minutes, followed by 50°C for 1 hour, then 70°C for 15 minutes.Note: cDNA may be stored at −20°C.
- To 1µl cDNA add 2.5µl 10× Platinum Taq PCR buffer, 1.2µl 50mM MgCl2, 0.5µl 10mM dNTPs, 0.25µl 25× SYBR Green, 0.2µl 25µM forward and reverse primer mix, 0.1µl Platinum Taq, and H2O to a total volume of 25µl.Note: 25× SYBR Green is prepared before hand by diluting 1:400 with H2O and stored at 4°C in the dark.
Run PCR plate at 95°C for 2 minutes, followed by 40 cycles of 95°C for 15 seconds, 55°C for 20 seconds, 72°C for 45 seconds.
A.3 Simple ChIP-seq Data Analysis Pipeline
Note: Commands for each step are italicized. Ensure all software has been successfully downloaded and installed including Condetri, SOAP2, and CSAR.
Note: Perform steps 1 and 3 as well as 7 and 8 for each genotype/sample.
Note: Each command contains file names that will change depending on user preference. This example uses the name “wt.fastq” for the file obtained from sequencing and “TAIR10_chr_all.fas” for the genome reference file downloaded from the TAIR website.
- Trim reads: perl condetri_v2.2.pl -fastq1=wt.fastq -prefix=wt -rmNNote: wt.fastq contains the reads and quality scores from sequencing. The prefix parameter “-prefix=” is used to name the output file and should be changed with each sample. In step1 the output file will be named wt_trim.fastq. The parameter “-rmN” removes N base calls from the beginning of reads.
Build Genome Index (only needs to be done once): ./2bwt-builder TAIR10_chr_all.fas
- Align Reads to Genome: ./soap -a wt_trim.fastq -D TAIR10_chr_all.fas.index -o wt.soapNote: wt_trim.fastq is the file containing the trimmed reads from step 1. wt.soap is the name of the output file for this step.
Repeat Steps 1 and 3 for each sample changing the names of the appropriate files.
Start R: R
- Load CSAR package: > library(CSAR)Note: The symbols > or + denotes a new line and are displayed by R.
- Load Mapped Reads for each genotype: > wt <- loadMappedReads(“wt.soap”, format = "SOAP", header = FALSE)Note: In R results from each command are saved into temporary variables using the “<-“ symbols. The results from step seven are saved into “wt” and these results are called upon in step eight.
- Extend reads to fragmentation length, eliminate PCR amplification errors, and count reads: > wt.nhits <- mappedReads2Nhits(wt, file = “wt.nhits”, chr = c("1", "2", "3", "4", "5"), chrL = c(30427671, 19698289, 23459830, 18585056, 26975502), w = 250L, considerStrand = "Sum", uniquelyMapped = TRUE)Note: Parameter w is the fragmentation length after sonication and artificially extends the reads to this length. considerStrand = “Sum” is used to count all the reads regardless of the strand they map to, but can be changed to “Minimum” if only reads with equal numbers on both strands are desired. uniquelyMapped is used to remove duplicate reads due to PCR amplification errors during library preparation. Each chromosome’s length is specified by the chrL parameter.
Repeat steps 7 and 8 with each sample changing the names “wt”, “wt.soap”, and “wt.nhits” to “mutant”, “mutant.soap”, and “mutant.nhits” respectively.
Compare genotypes and calculate enrichment scores: > score <-ChIPseqScore(mutant.nhits, wt.nhits)
Create windows of enrichment between genotypes (these windows will later be filtered based on false discovery rate (FDR)): > TotalPeaks <- sigWin(score)
- Run each of the following to perform permutations in order to calculate score cutoff based on FDR.
>dir.create(“permutations”) >setwd(“permutations”) > for (j in 1:20) { + permutatedWinScores(nn = j, mutant, wt, fileOutput = "perm", chr = c("1","2","3","4","5"), chrL = c(30427671, 19698289, 23459830, 18585056, 26975502), w = 250L, considerStrand = "Sum", uniquelyMapped = TRUE) + } >permscores <- getPermutatedWinScores(file = "perm", nn=1:20) >cutoff <- getThreshold(winscores=TotalPeaks$score, permutatedScores=permscores, FDR=.001) >setwd(“‥/”) - Use cutoff value to create list of significant enrichment windows: >FilteredPeaks <- TotalPeaks[TotalPeaks$score > cutoff$threshold,]Note: Step 12 uses a cutoff FDR of 0.001which is somewhat strict. Changing the FDR value in step 12 will change the sensitivity and specificity of peaks in the final list.
- Write file with list of peaks: >write.table(FilteredPeaks, sep="\t", file="wtvsmutant.txt", row.names=FALSE)Note: Exit R and examine the file using a word processor. The list of peaks is saved as “wtvsmutant.txt” and includes peaks where the FDR is less than 0.001. This list includes the coordinates of binding sites along with the position of the peak summit, the peak enrichment score, and the length of the binding region.
Appendix B: Equipment, Reagents and Buffers
B.1 Equipment
Vacuum chamber (ChIP and RIP), Isotemp Vacuum Oven Model 280A (Fisher Scientific), self-cleaning dry vacuum system (Welch)
Refrigerated centrifuge (ChIP and RIP), Thermo Scientific Sorvall Legend X1 Centrifuge, Eppendorf centrifuge 5424
Sonicator (ChIP and RIP), Fisher Scientific, Sonic Dismembrator Model 100
Rotator (ChIP and RIP), Thermo Scientific Labquake Tube Rotator
Magnetic separator (ChIP and RIP), Promega MagneSphere Technology Magnetic Separation
Stand
Real-Time Thermal Cycler (ChIP, RIP, and RT-PCR), Biorad CFX Connect Real-time System
B.2 Reagents
DTT (ChIP, RIP, and RT-PCR), Fisher Scientific, BP172-5
Formaldehyde 0.5% (ChIP and RIP), Sigma-Aldrich, F1635-500ML
Glycine 2M (ChIP and RIP), Fisher Scientific, G46-1
PMSF 100mM (ChIP and RIP), Sigma-Aldrich, P7626-25G
Plant Protease Inhibitor [Sigma] (ChIP and RIP), Sigma-Aldrich, P9599-5ML
Miracloth (ChIP and RIP), VWR, 80058-394
Dynabeads Protein A Magnetic Beads [Invitrogen] (ChIP and RIP), Invitrogen, 100-01D
Protein/Tag specific antibody (ChIP and RIP)
RNA-grade Proteinase K 20mg/ml [Invitrogen] (ChIP and RIP), Invitrogen, 25530-049
25:24:1 Phenol-chloroform-isoamyl alcohol pH 7.5 (ChIP), Fisher Scientific, BP1752I-100
25:24:1 Phenol-chloroform-isoamyl alcohol pH 4.3 (RIP), Fisher Scientific, BP1754I-100
24:1 Chloroform-isoamyl alcohol (ChIP), Fisher Scientific (chloroform: C298500, isoamyl alcohol: BP1150-500; mix 24:1)
GlycoBlue 15mg/ml [Ambion] (ChIP and RIP), Fisher Scientific, NC9567599
3M NaOAc pH 5.2 (ChIP and RIP), Fisher Scientific (NaOAc: BP333-500, acetic acid: AC14893-0025)
96% EtOH (ChIP and RIP), Decon Labs Inc., 2701
Turbo DNase 2U/µl [Ambion] (RIP and RT-PCR), Fisher Scientific, NC9075048
Ribolock RNase Inhibitor 40U/µl [Fermentas] (RIP and RT-PCR), Fisher Scientific, FEREO0384
RNeasy Plant Mini Kit [Qiagen] (RT-PCR), Qiagen, 74904
25mM EDTA pH8 (RT-PCR), Fisher Scientific (EDTA: BP120500, NaOH: BP359-500)
Random Primer 500ng/µl [Invitrogen] (RT-PCR), life technologies, 48190-011
dNTPs 10mM [Promega] (RT-PCR), Fisher Scientific, PRU1515
Superscript III Reverse Transcriptase 200U/µl, 5 × FS buffer, 0.1M DTT [Invitrogen] (RT-PCR), life technologies, 18080-044
Platinum Taq 5U/µl, 10× Platinum Taq PCR Buffer, 50mM MgCl2 [Invitrogen] (RT-PCR), life technologies, 10966-034
SYBR Green I Nucleic Acid Gel Stain - 10,000X concentrate in DMSO [Invitrogen] (RT-PCR), life technologies, S-7563
Chelex-100 (ChIP), Bio-Rad 142-1253
Nalgene Rapid Flow Sterile Disposable Filter Unit with PES Membrane [Thermo Scientific]
(ChIP/RIP), Fisher Scientific, 09-741-02
Ultra-pure water (miliQ)
B.3 Buffers
Honda Buffer
0.44M Sucrose, Fisher Scientific, BP220-212
1.25% Ficoll 400, Sigma-Aldrich, F2637-100G
2.5% Dextran T40, Sigma-Aldrich, D1662-100G
20mM HEPES, KOH pH7.4, Fisher Scientific (HEPES: BP310-100, KOH: P250-1)
10mM MgCl2, Fisher Scientific, M33-500
0.5% Triton X-100, Fisher Scientific, BP151-500
Prepare HEPES first in 350ml and set pH, then add other components.
Make 500ml and filter through 0.2 micron filter.
Store at 4°C.
Add the day of experiment:
5mM DTT, Fisher Scientific, BP1725
1mM PMSF, Sigma-Aldrich, P7626-25G
1% Plant Protease Inhibitors, Sigma-Aldrich, P9599-5ML
8U/ml Ribolock RNase Inhibitor (RIP only), Fermentas, FEREO0384
Binding/Washing Buffer (B/W)
150mM NaCl, Fisher Scientific, BP358-212
20mM Tris HCl pH8, Fisher Scientific (Tris: BP152-5, HCl: A144-500)
2mM EDTA pH8, Fisher Scientific (EDTA: BP120500, NaOH: BP359-500)
1% Triton X-100, Fisher Scientific, BP151-500
0.1% SDS, Fisher Scientific, BP166-500
Add water to 500ml and filter through 0.2 micron filter.
Store at 4°C.
Add the day of experiment:
1mM PMSF, Sigma-Aldrich, P7626-25G
40U/ml Ribolock RNase Inhibitor (RIP only), Fermentas, FEREO0384
Nuclei Lysis Buffer (NLB)
50mM Tris-HCl pH8, Fisher Scientific (Tris: BP152-5, HCl: A144-500)
10mM EDTA, Fisher Scientific (EDTA: BP120500, NaOH: BP359-500)
1% SDS, Fisher Scientific, BP166-500
Add the day of the experiment:
1mM PMSF, Sigma-Aldrich, P7626-25G 1%
Plant Protease Inhibitors, Sigma-Aldrich, P9599-5ML
160U/ml Ribolock RNase Inhibitor (RIP only), Fermentas, FEREO0384
ChIP-Dilution Buffer
Note: Tris-HCl and EDTA are added from filtered stock solutions.
1.1% Triton X-100, Fisher Scientific, BP151-500
1.2mM EDTA pH8, Fisher Scientific (EDTA: BP120500, NaOH: BP359-500)
16.7mM Tris-HCl pH8, Fisher Scientific (Tris: BP152-5, HCl: A144-500)
167mM NaCl, Fisher Scientific, BP358-212
350U/ml Ribolock RNase Inhibitor (RIP only), Fermentas, FERE00384
Store at 4°C for ease of use.
Elution Buffer
Note: Tris-HCl and EDTA are added from filtered stock solutions.
100mM Tris HCl pH8, Fisher Scientific (Tris: BP152-5, HCl: A144-500)
10mM EDTA, Fisher Scientific (EDTA: BP120500, NaOH: BP359-500)
1% SDS, Fisher Scientific, BP166-500
Add the day of the experiment:
400U/ml Ribolock RNase Inhibitor (RIP only) Fermentas, FERE00384
TE Buffer
10mM Tris-HCl pH8, Fisher Scientific (Tris: BP152-5, HCl: A144-500)
1mM EDTA, Fisher Scientific (EDTA: BP120500, NaOH: BP359-500)
Appendix C: List of Primers
IGN29: CGTTTGTTTATGTAGGGCGAAAG and TAAAACTTTTCCCGCCAACCA.
ACTIN2: GAGAGATTCAGATGCCCAGAAGTC and TGGATTCCAGCAGCTTCCA.
References
- 1.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wierzbicki AT. The role of long non-coding RNA in transcriptional gene silencing. Curr. Opin. Plant Biol. 2012:517–522. doi: 10.1016/j.pbi.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJM. RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 5.Zheng Q, Rowley MJ, Böhmdorfer G, Sandhu D, Gregory BD, Wierzbicki AT. RNA polymerase V targets transcriptional silencing components to promoters of protein-coding genes. Plant J. 2012 doi: 10.1111/tpj.12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Rendtlew Danielsen JM, Yang Y-G, Qi Y. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 2011;12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- 9.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Shami M, Pontier D, Lahmy S, Braun L, Picart C, Vega D, Hakimi M-A, Jacobsen SE, Cooke R, Lagrange T. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes & Development. 2007;21:2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haag JR, Ream TS, Marasco M, Nicora CD, Norbeck AD, Pasa-Tolic L, Pikaard CS. In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Mol. Cell. 2012;48:811–818. doi: 10.1016/j.molcel.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He X-J, Hsu Y-F, Zhu S, Wierzbicki AT, Pontes O, Pikaard CS, Liu H-L, Wang C-S, Jin H, Zhu J-K. An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rowley MJ, Avrutsky MI, Sifuentes CJ, Pereira L, Wierzbicki AT. Independent chromatin binding of ARGONAUTE4 and SPT5L/KTF1 mediates transcriptional gene silencing. PLoS Genet. 2011;7:e1002120. doi: 10.1371/journal.pgen.1002120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bies-Etheve N, Pontier D, Lahmy S, Picart C, Vega D, Cooke R, Lagrange T. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 2009;10:649–654. doi: 10.1038/embor.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu Y, Rowley MJ, Böhmdorfer G, Wierzbicki AT. A SWI/SNF Chromatin-Remodeling Complex Acts in Noncoding RNA-Mediated Transcriptional Silencing. Mol. Cell. 2013;49:1–12. doi: 10.1016/j.molcel.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wierzbicki AT, Cocklin R, Mayampurath A, Lister R, Rowley MJ, Gregory BD, Ecker JR, Tang H, Pikaard CS. Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes Dev. 2012;26:1825–1836. doi: 10.1101/gad.197772.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong X, Hale CJ, Law JA, Johnson LM, Feng S, Tu A, Jacobsen SE. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat. Struct. Mol. Biol. 2012;19:870–875. doi: 10.1038/nsmb.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- 19.Wong SS. Chemistry of protein conjugation and cross-linking. Boca Raton: CRC Press; 1991. [Google Scholar]
- 20.Nelson JD, Denisenko O, Sova P, Bomsztyk K. Fast chromatin immunoprecipitation assay. Nucleic Acids Res. 2006;34:e2. doi: 10.1093/nar/gnj004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 23.König J, Zarnack K, Luscombe NM, Ule J. Protein-RNA interactions: new genomic technologies and perspectives. Nat. Rev. Genet. 2011;13:77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- 24.Muiño JM, Kaufmann K, van Ham RC, Angenent GC, Krajewski P. ChIP-seq Analysis in R (CSAR): An R package for the statistical detection of protein-bound genomic regions. Plant Methods. 2011;7:11. doi: 10.1186/1746-4811-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li R, Yu C, Li Y, Lam T-W, Yiu S-M, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 26.Smeds L, Künstner A. ConDeTri--a content dependent read trimmer for Illumina data. PLoS ONE. 2011;6:e26314. doi: 10.1371/journal.pone.0026314. [DOI] [PMC free article] [PubMed] [Google Scholar]