Figure 3. In a phylogenetic analysis SbMATE2 is part of a clade containing MATE transporters for flavonoids and alkaloids.
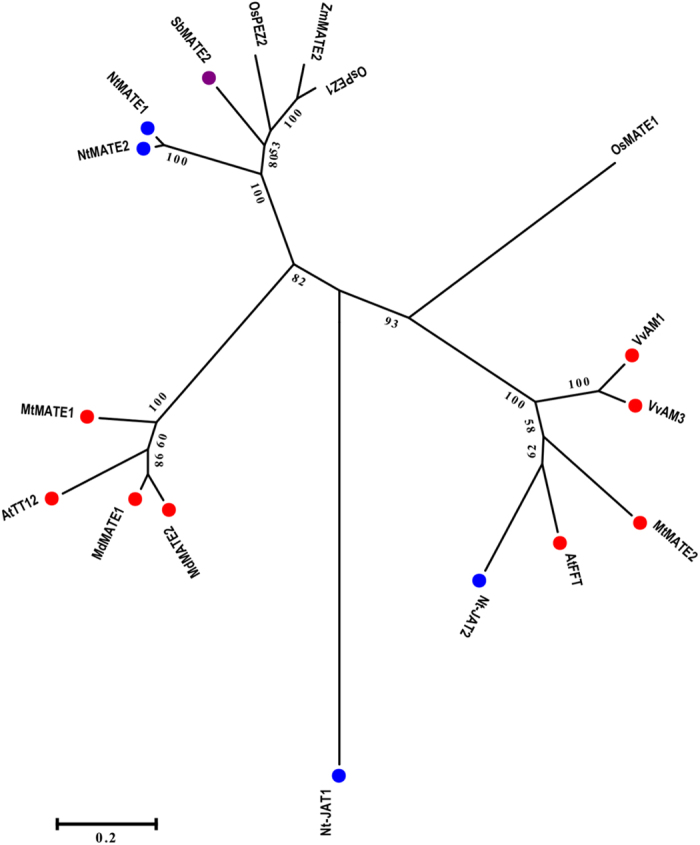
A molecular phylogenetic analysis was performed using the Maximum Likelihood method based on the Jones-Taylor-Thornton (JTT) matrix-based model for amino acid sequences. Branch lengths are measured in the number of substitutions per site, and positions containing gaps were eliminated. Bootstrap values (1000x) are indicated at branch points. Analyses were conducted using the MEGA5 software package47. The amino sequences used in the analysis were: SbMATE2 (Sorghum bicolor, Sobic.001G012600), OsPEZ1 (Oryza sativa, Os03g37490), OsPEZ2 (O. sativa, Os03g0572900), OsMATE1 (O. sativa, Os03g08900), TT12 (Arabidopsis thaliana, At3g59030), FFT (A. thaliana, At4g25640), MdMATE1 (Malus domestica, GU64954), MdMATE2 (M. domestica, GU064956), MtMATE1 (Medicago truncatula, FJ858726), MtMATE2 (M. truncatula, HM856605), VvAM1 (Vitis vinifera, Fj264202), VvAM3 (V. vinifera, FJ264203), NtMATE1 (Nicotiana tabacum, AB286961), NtMATE2 (N. tabacum, AB286962), Nt-JAT1 (N. tabacum, AM991692), Nt-JAT2 (N. tabacum, AB922128), and ZmMATE2 (Zea mays, FJ873684). Coloured circles represent the transported compound classes: red = flavonoids, blue = alkaloids, purple = hydroxynitrile glucosides.
