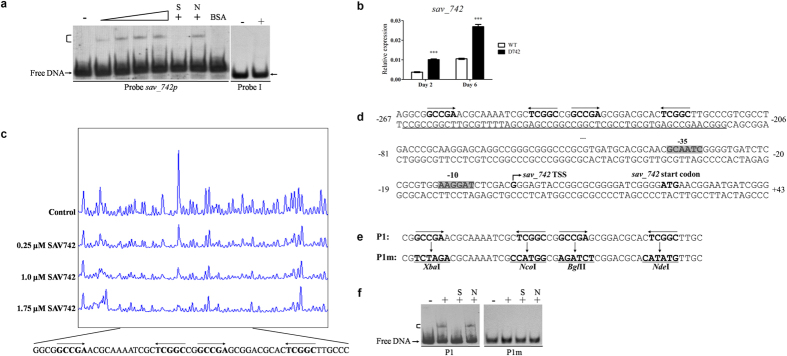
Figure 2. SAV742 directly represses its own gene.
(a) EMSAs of the interaction of His6-SAV742 with its own promoter region. 0.15 nM labeled probe was added to each reaction mixture. For specific (lane S) or nonspecific (lane N) competition assays, a 300-fold excess of unlabeled competitor DNA was used. 0.1% BSA and labeled nonspecific probe I were used as negative protein and probe controls, respectively. Lanes –: EMSAs without His6-SAV742. Lanes 2 to 5 contained 125, 250, 375, and 500 nM His6-SAV742, respectively. 500 nM His6-SAV742 was used for competition assays and probe I (Lanes +). Arrows: free probes. Bracket: SAV742-DNA complex. (b) qRT-PCR analysis of sav_742 transcription level in WT and D742 grown in FM-I for 2 or 6 days. Relative values were obtained using 16 S rRNA as internal reference. sav_742: 81-bp transcript amplified from the remaining sav_742 ORF in D742 with primers SD66A and SD66B. ***P < 0.001 (Student’s t-test). (c) Determination of SAV742-binding sites on its own promoter region by DNase I footprinting assay. Top fluorogram: control reaction without protein. Protection regions were acquired with increasing concentrations (0.25, 1.0, 1.75 μM) of His6-SAV742 protein. (d) Nucleotide sequences of sav_742 promoter region and SAV742-binding sites. Numbers indicate distance (nt) from sav_742 TSS. Bent arrow: sav_742 TSS. Straight arrows: inverted repeats. Shaded areas: putative -10 and -35 regions. Underlining: SAV742-binding site. (e) Mutational analysis of SAV742-binding sites. Each probe was 50-bp. Mutations were introduced into WT probe P1 to generate mutated probe P1m. Underlining: altered nucleotides. (f) EMSAs using WT probe P1 and mutated probe P1m. Each lane contained 0.15 nM labeled probe. A 300-fold excess of unlabeled competitor probe was used in competition assays. His6-SAV742 concentration: 500 nM.