Figure 5.
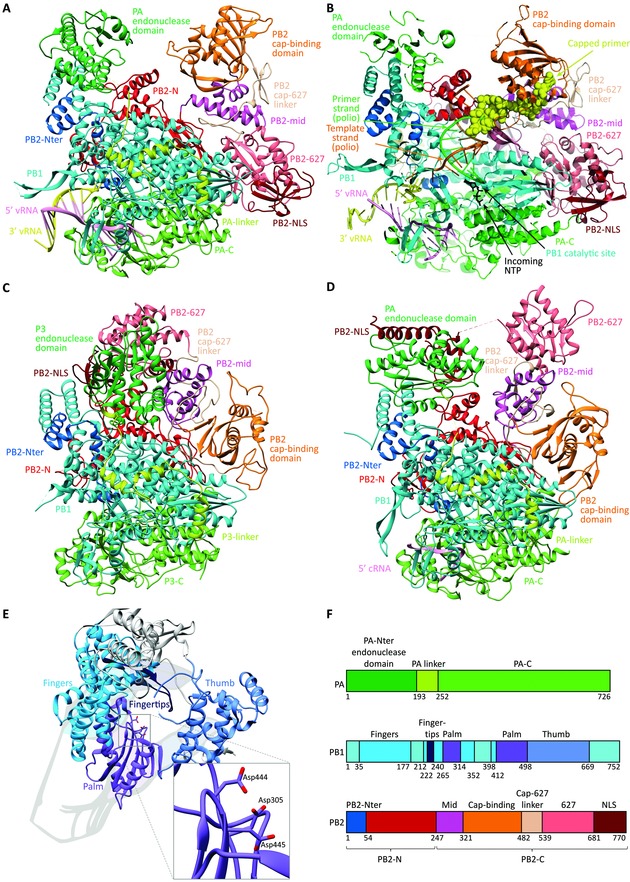
Comparison of the crystal structures of the heterotrimeric influenza polymerase complex containing full‐length PA, PB1, and PB2. The models are shown in the same orientation, and the same coloring was applied for the different subdomains. (A) Bat FluA polymerase with bound vRNA promoter [PDB: 4WSB].23 (B) Superposition model of the FluB polymerase crystal structure with a template–primer (orange–green) duplex and incoming NTP (black) (taken from a poliovirus polymerase crystal structure). The yellow spheres represent the capped primer bound to PB2, after cleavage by the PA endonuclease domain. This primer is now directed toward the PB1 catalytic cavity, where primer elongation occurs. (Adapted by permission from Macmillan Publishers Ltd: Nature, Reich et al.,24 copyright 2014.) (C) Influenza C polymerase (PDB: 5D98) in apo form.58 (D) FluB polymerase structure with bound cRNA 5′ end56 (PDB: 5EPI). (E) Domain arrangement of FluB PB1, illustrating the right‐hand‐like polymerase fold. The inset shows a closeup of the PB1 catalytic residues, which coordinate two divalent metal ions (not shown). (F) Subdomain names and color scheme as applied in panels A–E, based on the FluB polymerase numbering. For clarity, the PB1 subunit is colored uniformly in cyan in panels A–D, while its different subdomains are differentiated in panel E.
