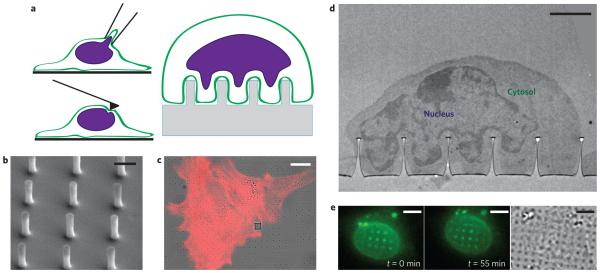
Figure 1. The nuclear envelope deforms around nanopillars, creating a non-invasive platform for studying in situ nuclear deformation.
a, Illustrations of techniques for studying nuclear mechanics. Anticlockwise from top left: micropipette aspiration, atomic force microscopy and nanopillar arrays. b, SEM image of nanopillar array with 75 nm radius, 2 μm pitch and 1.4 μm height. Scale bar, 1 μm. c, Overlay of differential interference contrast (DIC) and fluorescence images of 3T3 cells cultured on a nanopillar array. The red colour shows actin staining, and nanopillars are visible as dark spots in the bright-field image. The grey square in the DIC image is an alignment marker used to locate the nanopillar arrays. Scale bar, 10 μm. d, TEM image showing the nucleus deformed around a nanopillar array with the cytosol visible between the nuclear envelope and the cell membrane. Scale bar, 2 μm. e, Fluorescence (left and middle, different time points) and DIC (right) images of a live 3T3 cell transfected with GFP-Sun2. Nuclear deformation is evident as bright fluorescent spots around the nanopillars and changes as the cell migrates. See Supplementary Movies 1 and 2 for complete time lapse of nuclear deformation during cell migration. Scale bars, 5 μm.