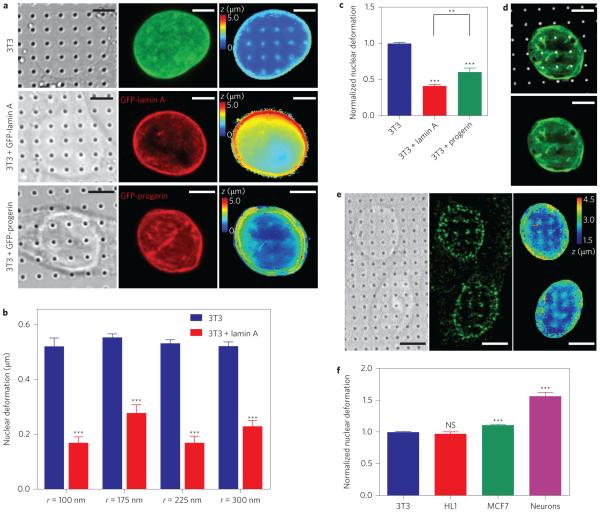
Figure 3. Nanopillar-induced nuclear deformation shows a strong dependence on nuclear stiffness.
a, Reconstructed surfaces of the nuclear envelope reveal differences in the deformability of an untransfected 3T3 cell (top), a GFP-lamin A-transfected 3T3 cell (middle) and a GFP-progerin-transfected 3T3 cell (bottom). The locations of the nanopillars were identified by DIC imaging (left column) and the nuclear envelope was visualized by fixing the cells and immunostaining with anti-lamin A (untransfected cells, green) or anti-GFP (transfected cells, red) antibodies (middle column). The surface of the nuclear envelope (right column, false-coloured in z position) is visibly deformed at nanopillars in the untransfected cell but less so in GFP-progerin-transfected and GFP-lamin A-transfected cells. b, Average nuclear deformation, consistently showing less deformation for lamin A-transfected cells than for untransfected 3T3 cells for all nanopillar geometries tested. Nanopillars with four different radii are shown, but with the same pitch of 3 μm and height of 1.4 μm. c, When normalized to untransfected 3T3 cells on the same geometry, 3T3 cells transfected with lamin A and progerin cells show much less deformation than untransfected cells. Interestingly, progerin-transfected cells show more deformation than lamin A-transfected cells. d, GFP-progerin-transfected 3T3 cell shows characteristic folds in the nuclear lamina (green). These folds align with nanopillar locations (false-coloured white) at the basal nuclear envelope. e, Representative images of hippocampal neurons show significant nuclear deformation on nanopillars. Left: DIC image showing pillar locations. Middle: Immunostaining image with anti-nuclear pore complex illustrating nuclear envelope. Right: Reconstructed z surface of nuclear envelope (false coloured in z) showing that the nuclear envelope is deformed at nanopillar locations. f, Average nuclear deformation in different cell types, normalized to 3T3 cells on the same geometry. HL1 and 3T3 cells are equally deformable, while MCF7 cells show more deformation. Neurons show significantly more deformation than any of the other cell types. All measurements were performed in triplicate. The numbers of nanopillars and nuclei included in each sample are provided in Supplementary Table 1. Comparisons were analysed with two-tailed, unequal variance student t-tests, corrected for multiple comparisons where necessary (c). **P < 0.01, ***P < 0.001. NS, no significant difference. Error bars indicate standard error of the mean. Scale bars, 5 μm.