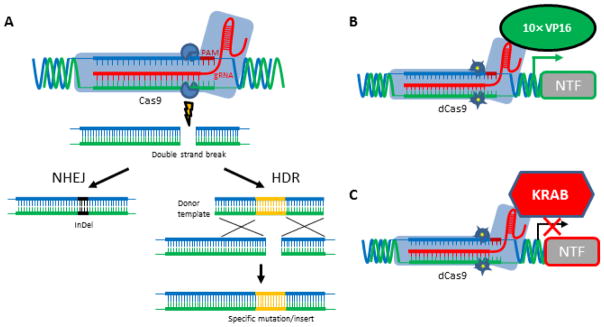
Figure 2.
CRISPR-Cas9 for controlling gene expression. (A) Binding of 20 nucleotides (nt) within the guide RNA (gRNA) to the complementary genomic DNA sequence preceding the 3 nt protospacer-associated motif (PAM) targets Cas9 to induce a site-specific double-strand break that is repaired by non-homologous end joining (NHEJ) or homology-directed repair (HDR). NHEJ frequently produces small insertions or deletions (InDels) that can lead to frameshift mutations in the target gene. In the presence of donor DNA template HDR leads to the insertion (knock-in) of specific DNA sequence to the target locus. (B) Activation or (C) inhibition of target gene transcription can be achieved by fusing the “dead” Cas9 (dCas9) protein carrying two point mutations blocking nuclease activity of Cas9 to VP16 transcriptional activator domains (10× VP16) or to a repressive chromatin modifier domain (KRAB), respectively. Images modified from (Charpentier and Doudna, 2013; Gilbert et al., 2014).