Figure 3.
YY1 binding to ECR47 enhancer is disrupted by rs2271338 risk allele.
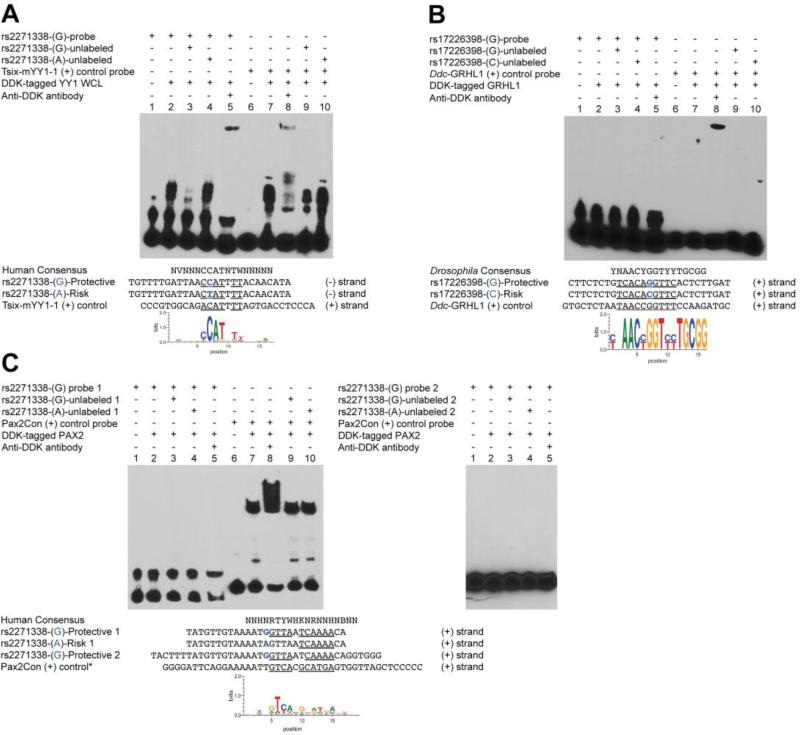
(A) A biotin-labeled DNA fragment containing rs2271338 ADHD protective allele was incubated with a HEK293 whole cell lysate (WCL) expressing DDK (FLAG)-tagged human YY1 (lanes 2-5 from left to right). Mobility shift in lane 2 indicates protein binding to the probe, which was abrogated by incubation with molar excess of the unlabeled fragment (lane 3), but not of an unlabeled fragment containing the rs2271338 risk allele (lane 4). Higher molecular weight shift with the addition of anti-DDK antibody identifies YY1 as the binding factor (lane 5). Lanes 6-10 correspond to a known YY1 binding sequence used as positive control (87). Lanes 7-10 show protein binding to the control probe, with the anti-DDK antibody producing a similar supershift thus confirming YY1 identity (lane 8). Molar excess of the unlabeled ECR47 protective fragment was capable of reducing protein binding (lane 9), but the risk fragment was not (lane 10).
No binding was detected for GRHL1 (B) or PAX2 (C) transcription factors. (B) Lanes 1-5, biotin-labeled fragment containing the rs17226398 protective allele; lanes 6-10, GRHL1 positive control sequence (89). Interestingly, no shift was observed in the positive control except when the antibody was added (lane 8). Apparently, the addition of the antibody stabilizes GRHL1-DNA interaction, which otherwise is labile under the experimental conditions used. (C) Left panel, lanes 1-5, same DNA fragment used for YY1 in A; lanes 6-10, PAX2 positive control sequence (90). Nether the protective nor the risk ECR47 fragments affected PAX2 binding to the positive control probe (lanes 9 and 10). The right panel shows the results for a longer ECR47 probe (Protective 2) extending to the 3’ end of the sequence. PAX2 still did not bind to DNA.
Position weight matrices were taken from MotifMap for human hg19 (YY1, PAX2) and Drosophila dm3 (GRHL1) assemblies (91).