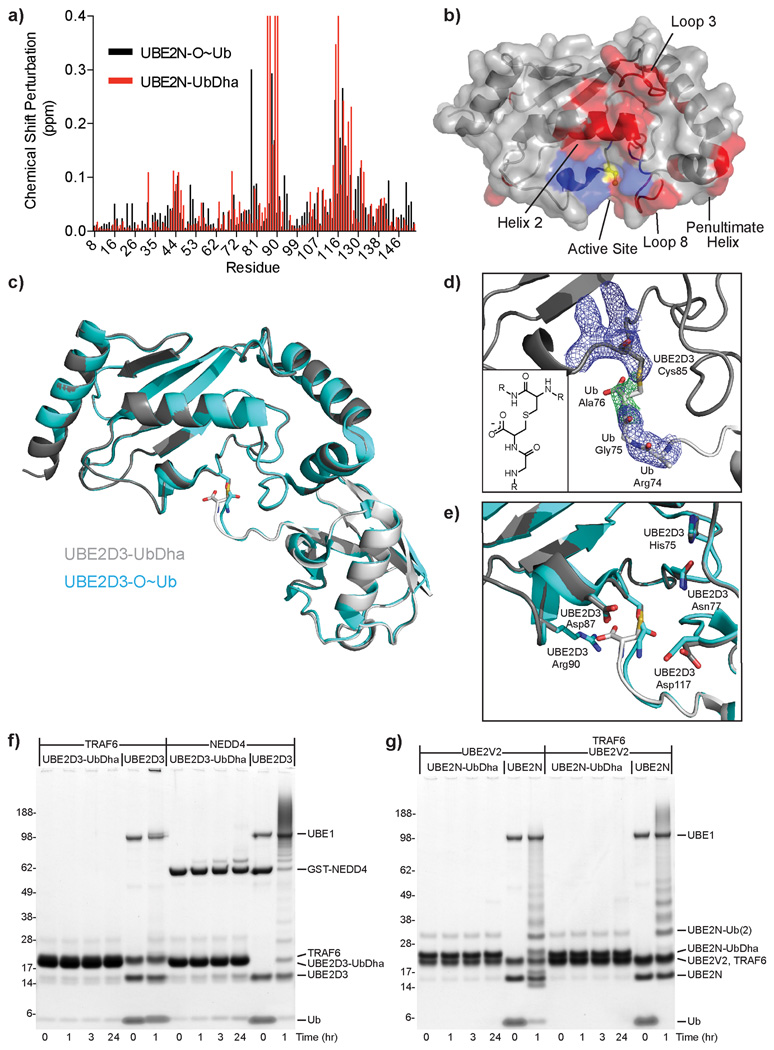
Figure 3. Structural studies of thioether-linked E2-Ub adducts.
a) UBE2N chemical shift perturbations upon Ub activation for oxyester (black) and thioether (red) linkages. Resonances perturbed beyond facile re-assignment were plotted using maximum perturbation value. b) Changes (from a) mapped onto the UBE2N structure (PDB 1J7D) (similarities in red; differences in blue). Active site Cys is colored yellow. c) Superposition of thioether-linked UBE2D3-UbDha conjugate (gray) over the oxyester-linked form (cyan, PDB 3UGB). d) Simulated annealing omit map of electron density surrounding the thioether linkage. 2|Fo|-|Fc| electron density (blue) contoured at 1σ, |Fo|-|Fc| density (green) contoured at 3σ. Inlay: diagram illustrating the thioether linkage. e) Overlay of E2 active site residues in thioether (gray) and oxyester (cyan) structures. Stability of the thioether-linked UBE2D3- and UBE2N-UbDHA adducts, incubated with f) RING E3 ligase TRAF6 or the HECT E3 ligase NEDD4, or g) accessory E2-variant UBE2V2, alone or in combination with the RING E3 ligase TRAF6.