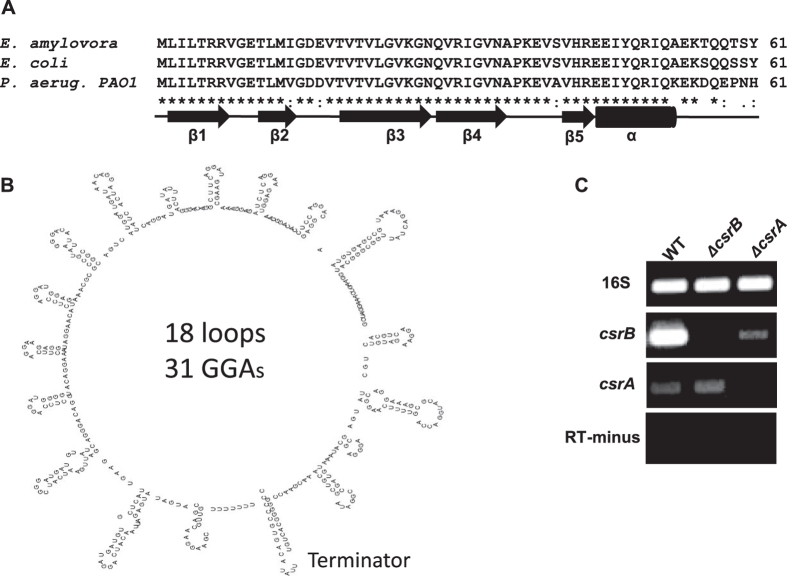
Figure 1. Alignment of deduced amino acids of CsrA and secondary structure of csrB from Erwinia amylovora.
(A) Alignment of deduced amino acids of CsrA (accession # CBJ47311 and NP_417176) from E. amylovora and Escherichia coli; and RsmA (accession #AAG04294) from Pseudomonas aeruginosa PAO1, and schematic map of the secondary structure of CsrA based on Schubert et al.56. Identical residues (*), conserved (:) and semi-conserved (.) substitutions are shown as underneath symbols. (B) Predicted secondary structure of csrB sRNA using the mfold program; (C) Gene expression of csrB and csrA by semi-quantitative PCR in wild type and the csrB and csrA mutants in MBMA medium. RT-minus indicates that no mRNA was added.