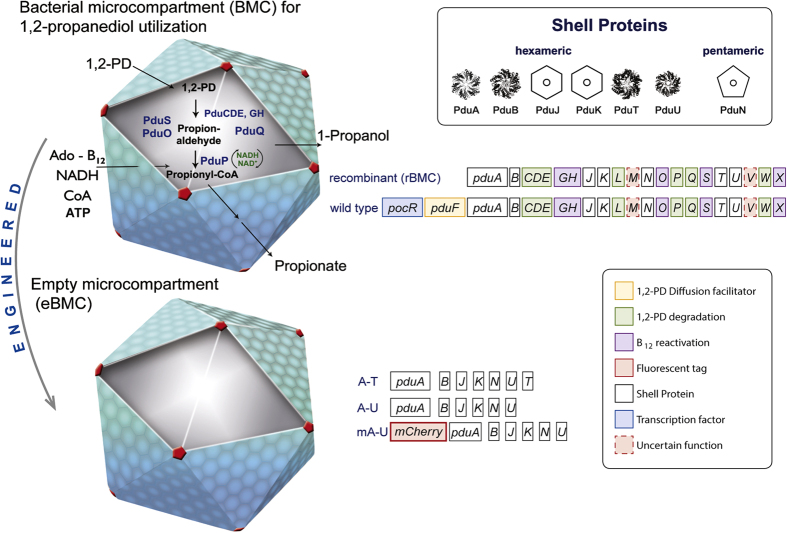
Figure 1. Schematic illustration of the Pdu shell and shell proteins associated with the wtBMC, rBMC and eBMC variants (A-U, mA-U and A-T).
Shell Proteins: The protein structures for PduA, PduB, PduT and PduU are shown, revealing the central pores within the hexameric or pseudohexameric symmetry. Hexameric shell proteins form the facets of the structure. The pentameric vertex protein (PduN) is thought to take up the vertex position of the structure. Bacterial microcompartment (BMC) for 1,2-propanediol utilization: wtBMCs and rBMCs incorporate many of the enzymes necessary for 1,2-PD degradation as well as enzymes for adenosylcobalamin reactivation. The rBMC plasmid lacks the genes encoding for the diffusion facilitator protein (yellow, pduF) and the transcription factor (cyano, pocR). Substrate (1,2-PD) and cofactors (ATP, CoA, NAD+ and B12) are thought to diffuse through the gated pores in the shell protein. Empty microcompartment (eBMC): Three different shell protein constructs consisting of shell protein genes (A-T, A-U and mA-U) can be produced in E. coli (Supplementary). A-T is the basic eBMC that houses all the shell proteins (PduA, B, B’, J, K. N. U, T). A-U is composed of six Pdu shell protein, PduA, B, B’, J, K, N, U. The mA-U construct has a C-terminal fusion of mCherry (red, fluorescent tag) to PduA and thus consists of mCherry-PduA, B, B’, J, K, N, U.