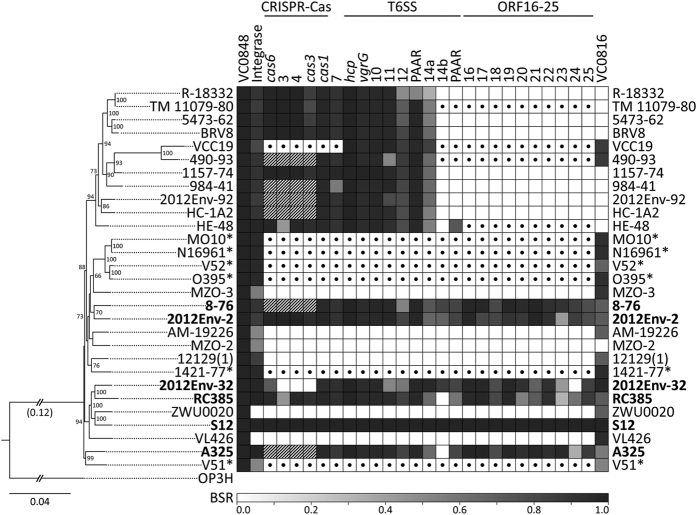
Figure 2. Presence/absence of the GIVchS12 ORFs in various Vibrio cholerae strains.
The heat map shows the BLAST score ratio (BSR) against the GIVchS12 reference (each column is an ORF). The gradient bar shows the BSR values and their corresponding colours; white indicates the absence of the ORF. Only BSR values of at least 0.3 were included. Strains with similar or complete GIVchS12 are indicated in bold. Striped boxes indicate the presence of a CRISPR-Cas module different from GIVchS12; dotted boxes indicate the presence of other ORFs in those regions different from GIVchS12. Strains with the VPI-1 similar to N16961 are indicated with *. The phylogenetic relationship of the V. cholerae strains is shown on the left of the heat map with Vibrio metoecus OP3H as outgroup. The maximum likelihood phylogenetic tree was constructed from the core alignment of whole genomes (≈2.5 mb) and using the GTR gamma substitution model with 100 bootstrap replicates (indicated on the tree nodes). The scale bar represents nucleotide substitutions per site. Shortened branch lengths, approximately three times the scale bar (0.12), are indicated.