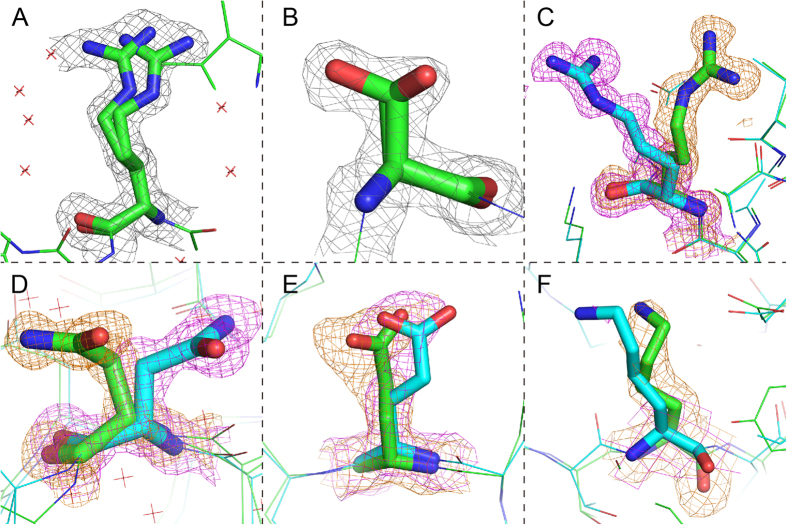
Figure 1. Side-chain conformational variation examples.
(A) Arg6 in plu4264 protein (4mv2) structure has two alternate locations, resolution 1.35 Å. State A has 0.45 occupancy, while state B has 0.55 occupancy. (B) Ser141 in 4b1y chain B includes two discrete alternate locations of 0.66 and 0.33 occupancies. (C) Arg112 in lysozyme C protein (3wl2, resolution 0.96 Å) chain A (green) and B (blue). Sequence identity of the two chains is 100%, while TMscore is >0.98. Electron density in both the structures are clear, while occupancies of all the atoms equal to 1. (D) Asn44 in 2vb1 (green, in chain A, resolution 0.65 Å) and 1iee (cyan, in chain A, resolution 0.94 Å). Both the chains are lysozyme C protein structures in Gallus gallus sharing 100% sequence identity and 0.97 TMscore, solved by different labs. Electron density map of 2vb1 is shown in green and 1iee is shown in blue. Both the residues have clear electron density for all the atoms. (E) Glu81 in 4pj2 (green, in chain A, resolution 1.24 Å) and 3od9 (cyan, in chain A, resolution 1.41 Å). Both the chains are periplasmic lysozyme inhibitor of I-type lysozyme structures in Aeromonas hydrophila sharing 100% sequence identity and >0.97 TMscore, solved by the same lab. Electron density map of 4pj2 is shown in green and 3od9 is shown in blue. The residue of 4pj2 has clear electron density for all the atoms, while not all of the atoms have clear electron density in 3od9. (F) Lys95 in 2pa8 (green, in chain D, resolution 1.76 Å) and 2pmz (cyan, in chain (D), resolution 3.4 Å). Both the chains are DNA-directed RNA polymerase subunit D structures in Sulfolobus solfataricus sharing 100% sequence identity and >0.91 TMscore, solved by the same lab. Electron density map of 2pa8 is shown in green and 2 pmz is shown in blue. The residue of 2pa8 has clear electron density for all the atoms, while none of the side-chain atoms has clear electron density.