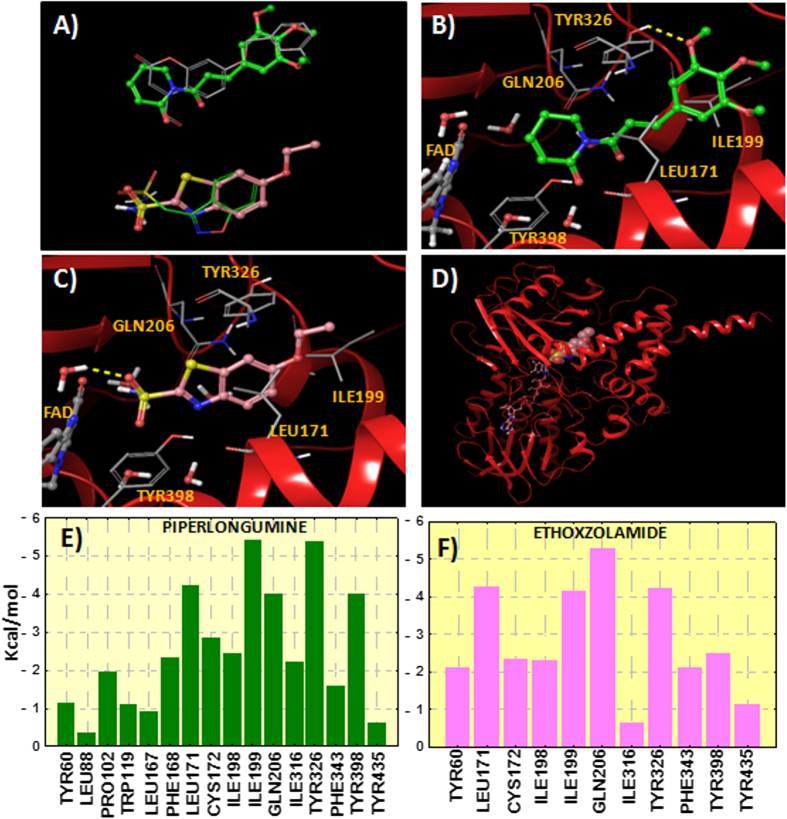
Figure 6.
(A) Comparison between the theoretical poses determined for piperlongumine (green carbons) and ethoxzolamide (pink carbons) and the co-crystallized inhibitors c17 (2V60) and zonisamide (3PO7) inside the hMAO-B. (B) Binding mode for piperlongumine extracted from the hMAO-B docking. Hydrogen bond is represented in yellow color. Protein ribbons are partially omitted for clarity. (C) Binding mode determined for ethoxzolamide inside the hMAO-B. (D) Perspective of the whole hMAO-B enzyme with ethoxzolamide docked to the protein (FAD cofactor in stick representation and ethoxzolamide in CPK). (E) Residue interaction energy scores between hMAO-B and piperlongumine. (F) Residue interactions with ethoxzolamide. Contribution of the residues in a distance of 4 Å from the ligands is plotted. Interaction energy is calculated as the sum of the contributions of Coulomb, van der Waals and hydrogen bonding.