Fig. 7.
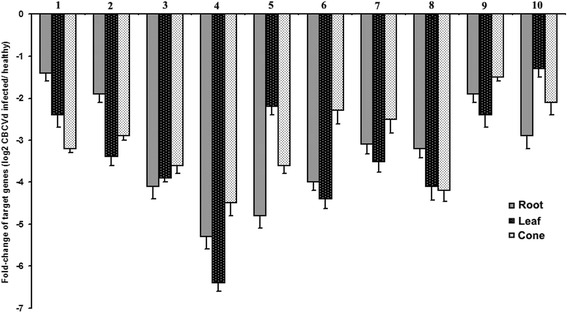
Differential expression profile of selected miRNA target genes: The relative gene expression was evaluated using the comparative Cq method taking Hl-GAPDH (GenBank Accession No. ES437736) as the reference gene. The log2 values represent the ratio of the expression level of target genes in CBCVd-infected samples relative to healthy samples in root, leaf and cone. The predicted target genes used in this analysis were (1) Squamosa promoter-binding protein 15 (hop-miR156 Target); (2) GAMYB Transcriptional factor (hop-miR159c Target); (3) NAC domain protein NAC1 (hop-miR164a Target); (4) Cysteine protease (hop-miR167a-5p Target); (5) mediator of RNA pol II subunit 26b (hop-miR167c-5p Target); (6) GRAS family TF (hop-miR171b Target); (7) pre-mRNA-splicing factor RNA helicase PRP1 (hop-miR395a Target); (8) Glycerol-3-phosphate dehydrogenase (hop-miR827a Target); (9) disease resistance protein RPM1-like (hop-miR02 Target); (10) cellulose synthase A catalytic subunit 4 (hop-miR41 Target). Target genes with a significant change of (P < 0.05 and log2 Ratio > 1) were considered to be differentially expressed. Values are given as mean ± SD of three experiments in each group
