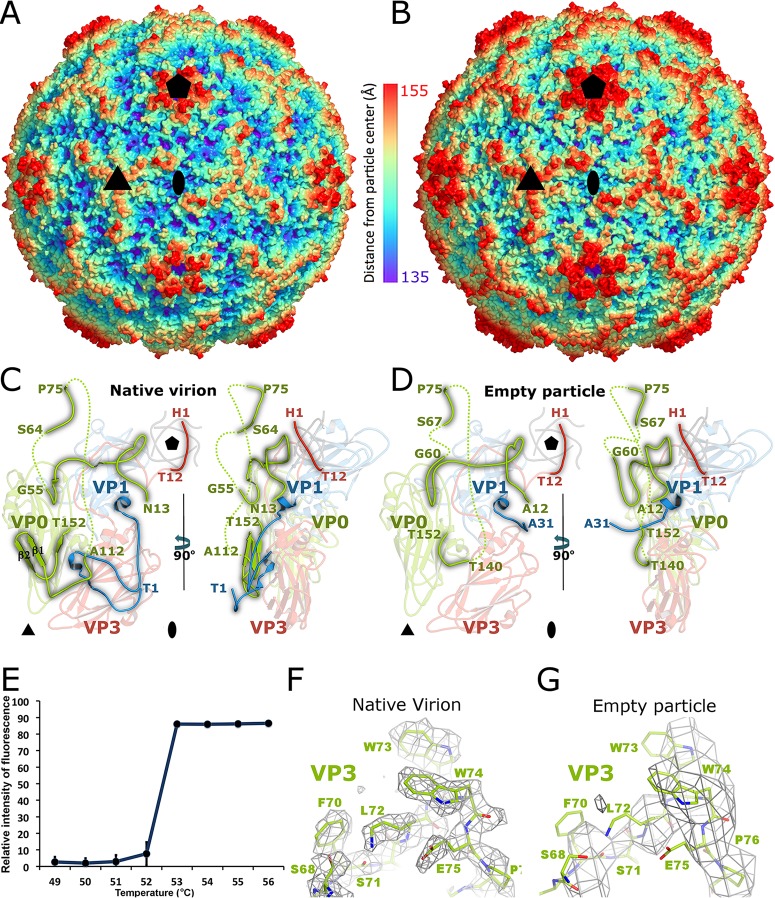
FIG 2.
Changes of AiV-1 capsid associated with genome release. (A and B) Surface rendering of AiV-1 virion (A) and empty particle (B) rainbow-colored based on distance from the particle center. The positions of selected 5-fold, 3-fold, and 2-fold icosahedral symmetry axes are indicated with pentagons, triangles, and ovals, respectively. (C and D) Icosahedral asymmetric unit of native AiV-1 virion (C) and the empty particle (D). VP1, VP0, and VP3 are shown in blue, green, and red, respectively. N-terminal arms of the capsid proteins are highlighted in brighter colors. The positions of the 5-fold-symmetry-related N termini of VP3 subunits are shown in gray. Dashed lines indicate the putative positions of the unstructured chains. The positions of 5-fold, 3-fold, and 2-fold icosahedral symmetry axes are indicated with pentagons, triangles, and ovals, respectively. (E) A Sybr green fluorescence assay was performed to measure the stability of AiV-1 virions. AiV-1 virions were mixed with Sybr green dye II and heated to the indicated temperatures (x axis). The fluorescence signal increases as the dye binds to RNA that is released from thermally destabilized particles. Error bars indicate the standard deviations of the measurements. Please see Materials and Methods for details. (E and F) Examples of electron densities of AiV-1 virion at a resolution of 2.1 Å (E) and empty particle at the resolution of 4.2 Å (F). The maps are contoured at 1.5σ.