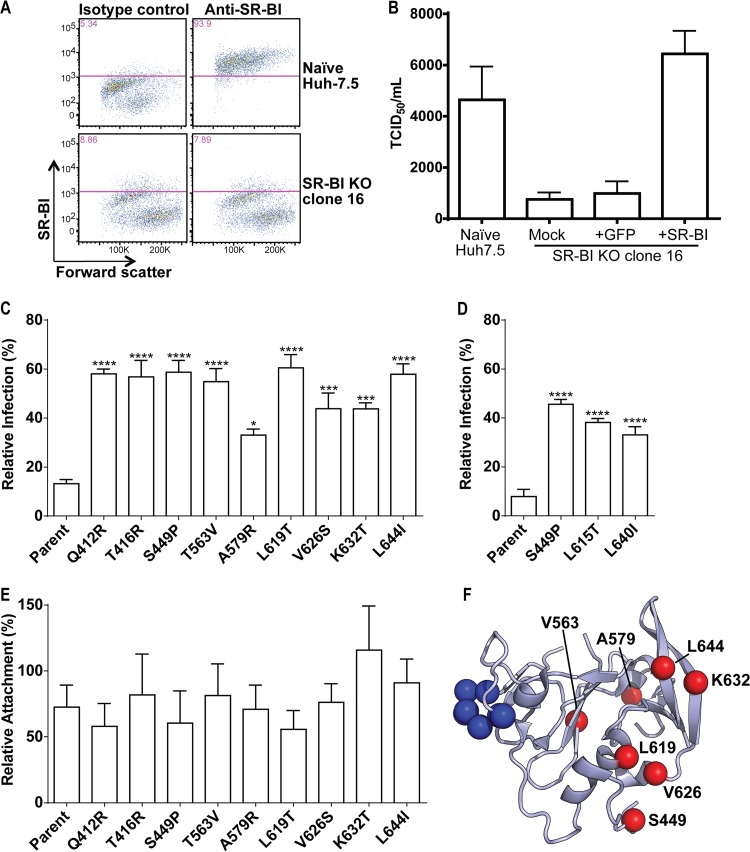
FIG 7.
Generation of SR-BI-deficient Huh7.5 cells. (A) Parental Huh7.5 cells or Huh7.5 SR-BIKO clone cell populations were stained with isotype control or SR-BI-specific antibodies and analyzed by flow cytometry. (B) The relative titers of a single stock of HCV were determined by limiting dilution assay on parental Huh7.5 cells or Huh7.5 SR-BIKO clonal cells either not expressing a transgene (mock) or transduced with lentiviral particles to express GFP or SR-BI. Values represent the mean TCID50/ml, and the error bars indicate SEM from three independent assays. (C) SR-BIKO Huh7.5 cells stably expressing GFP or GFP and SR-BI were infected with parental or variant virus. The relative infection of GFP-only cells compared to GFP- and SR-BI-coexpressing cells is shown. Error bars represent SEM. Asterisks indicate differences that are statistically significant (*, P < 0.05; ***, P < 0.001; ****, P < 0.0001). The results are averages from three independent experiments performed in quadruplicate. (D) SR-BIKO Huh7.5 cells stably expressing GFP or GFP and SR-BI were infected as described for panel C using parental H77/JFH-1 and H77/JFH-1 variant viruses bearing corresponding mutations to JFH-1 S449P, L619T, and L644: S449P, L615T, and L640L, respectively. (E) SR-BIKO Huh7.5 cells stably expressing GFP or GFP and SRBI were incubated with parental or variant virus for 3 h on ice. Cells were washed extensively, cellular RNA was harvested, and HCV genome copies were quantified by qRT-PCR. The relative attachment to GFP-only cells compared to GFP- and SR-BI-coexpressing cells is shown. Error bars represent SEM. No statistically significant differences were observed. The results are averages from three independent experiments performed in duplicate. (F) Ribbon diagram of the E2 core structure (PDB entry 4MWF) (19) illustrating the locations of the adaptive mutations identified in this study (red spheres) and residues essential for CD81 binding (blue spheres; amino acid positions 529 to 535). Q412 and T416 were excluded as they fall within a disordered region.