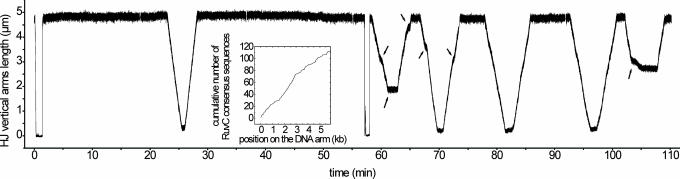
Fig. 2.
Influence of RuvB concentration. The experiment was performed by using a multi-ss construction. All along this experiment, the same DNA molecule was followed, and a constant load force of 0.73 pN was applied. In the first part of the experiment (from 0 to 58 min), 100 nM RuvA and 150 nM RuvB were injected in the sample (see the downfall near 0 min). In the second part (from 58 to 110 min), 100 nM RuvA and 670 nM RuvB were injected (see the downfall near 58 min). Whereas in the presence of 150 nM RuvB only one back-and-forth migration occurs during ≈1 h and with a nearly constant speed, in the presence of 670 nM RuvB the frequency of back-and-forth migrations is clearly higher and the migrations are less regular, because one can perceive migration inversions (arrows) and more frequent variations in speed (arrows). (Inset) Repartition of the RuvC consensus sequence 5′-(A/T)TT(G/C)-3′ along the vertical DNA arm sequence (a high slope corresponds to a high density of RuvC consensus sequences).