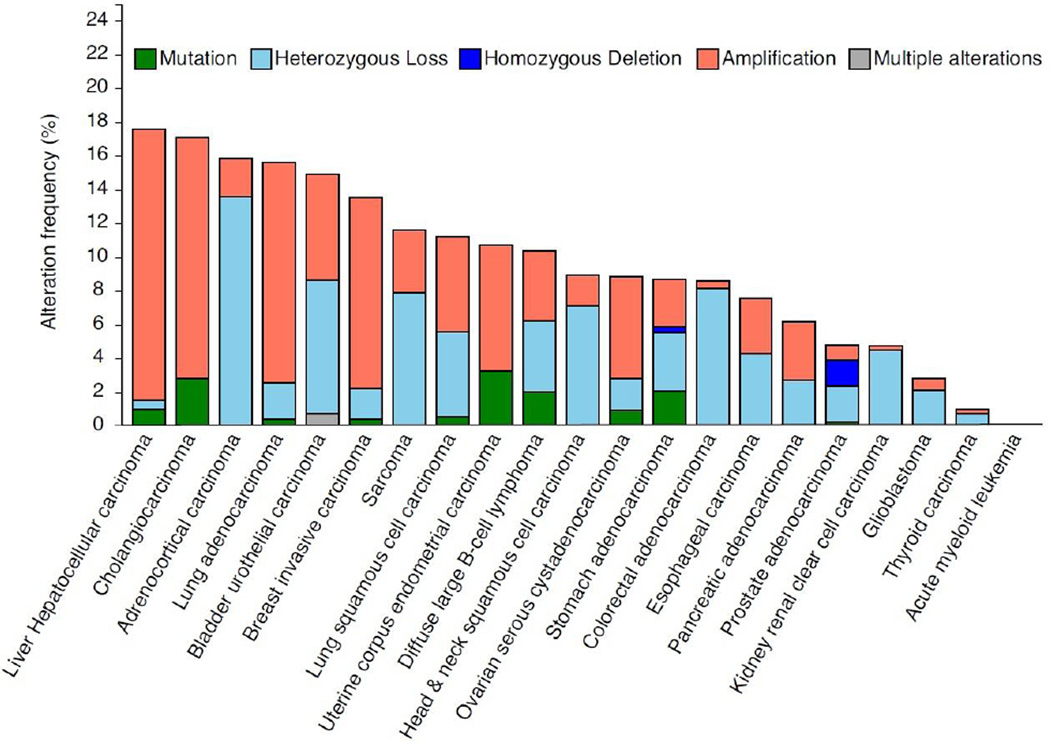
Figure 3. Frequency of mutations and copy-number alterations in the LMNA gene in human cancers.
The results shown here are based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/) and analyzed via cBioPortal (http://www.cbioportal.org/) (Cerami et al., 2012; Gao et al., 2013) on April 10, 2016. These results should therefore not be viewed as original data, but as reference to existing publicly available resources. In the analysis, shallow loss determined by copy-numberanalysis is referred to here as “heterozygous deletion” and deep loss is referred to as “homozygous deletion”. Copy number variations make up the majority of genomic changes in the LMNA gene in cancer, whereas mutations are rare. It remains to be determined whether LMNA genomic alterations correlate closely with protein levels in cancer, and which transcriptional and/or post-translational mechanisms are important contributors to altered lamin A/C expression in cancer.