Abstract
BACKGROUND: HMGA1 is a non-histone nuclear protein that regulates cellular proliferation, invasion and apoptosis and is overexpressed in many carcinomas. In this study we sought to explore the expression of HMGA1 in HCCs and cirrhotic tissues, and its effect in in vitro models. METHODS: We evaluated HMGA1 expression using gene expression microarrays (59 HCCs, of which 37 were matched with their corresponding cirrhotic tissue and 5 normal liver donors) and tissue microarray (192 HCCs, 108 cirrhotic tissues and 79 normal liver samples). HMGA1 expression was correlated with clinicopathologic features and patient outcome. Four liver cancer cell lines with stable induced or knockdown expression of HMGA1 were characterized using in vitro assays, including proliferation, migration and anchorage-independent growth. RESULTS: HMGA1 expression increased monotonically from normal liver tissues to cirrhotic tissue to HCC (P < .01) and was associated with Edmondson grade (P < .01). Overall, 51% and 42% of HCCs and cirrhotic tissues expressed HMGA1, respectively. Patients with HMGA1-positive HCCs had earlier disease progression and worse overall survival. Forced expression of HMGA1 in liver cancer models resulted in increased cell growth and migration, and vice versa. Soft agar assay showed that forced expression of HMGA1 led to increased foci formation, suggesting an oncogenic role of HMGA1 in hepatocarcinogenesis. CONCLUSIONS: HMGA1 is frequently expressed in cirrhotic tissues and HCCs and its expression is associated with high Edmondson grade and worse prognosis in HCC. Our results suggest that HMGA1 may act as oncogenic driver of progression, implicating it in tumor growth and migration potential in liver carcinogenesis.
Introduction
HMGA1 is a non-histone nuclear protein involved in cell cycle-related chromosomal changes, genetic recombination, DNA replication and repair, apoptosis, and molecular chaperoning [1], [2], [3], [4]. HMGA1 functions as an architectural transcriptional factor, as it regulates its target genes and microRNAs by direct DNA binding, forming transcriptional complexes and altering the conformation of transcription factors and chromatin structure [5], [6], [7]. HMGA1 is generally not expressed in adult tissues but is enriched in human embryonic and hematopoietic stem cells [8].
HMGA1 was first associated with the neoplastic phenotype in rat thyroid transformed cells [9] and has since been shown to lead to neoplastic transformation [3]. Of its many roles, HMGA1 negatively regulates TP53 [10] and promotes an undifferentiated pluripotent stem-like cell state through the induction of SOX2, LIN28 and cMYC [11]. HMGA1 also directly activates genes involved in tumor growth, migration, invasion, resistance to drug-induced cell death and epithelial-mesenchymal transition in cancer cells [12], [13], [14], [15]. Indeed, HMGA1 overexpression has been reported in carcinomas of the colon, breast, pancreas, ovary, lung, esophagus and testis [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], and it correlated with advanced stage, the presence of distant metastases and poor survival in colorectal carcinomas [16], [17]. Furthermore, HMGA1 expression levels have been found to increase progressively from no expression in normal breast tissue, to moderate expression in hyperplastic lesions to strong overexpression in ductal carcinomas [18], and to increase from weakly expressed in ovarian carcinomas with low invasive potential to be highly expressed in invasive carcinomas [21].
The HMGA1 locus (6p21.3) is gained in around 40% of hepatocellular cancers (HCCs) [26], and an early study suggested that HMGA1 is expressed in 30% of primary HCC on the mRNA level and 13% on the protein level [27]. Furthermore, HMGA1 mRNA expression was found to correlate with Edmondson grade and worse prognosis [27]. The functional significance of HMGA1 overexpression, however, has not been assessed. In this study, we evaluated HMGA1 mRNA expression levels in a cohort of HCC needle biopsies matched with their corresponding cirrhotic tissues and normal liver donors by gene expression microarrays and quantitative real-time PCR (qRT-PCR). Using tissue microarray (TMA) technology, we further corroborated our results at the protein level in a large independent collection of 379 specimens including normal liver, cirrhotic and HCC tissues. Finally, we showed that HMGA1 overexpression promoted tumor growth and migration potential in liver carcinogenesis.
Materials and Methods
Ethics
The study has been approved by the Institutional Review Board of the Institute of Pathology, University Hospital, Basel and the Ethics Committee of Nordwest/Central Switzerland (EKNZ).
Re-analysis of Transcriptomic Profiling Data
HMGA1 expression was evaluated in 59 HCC needle biopsies, 37 of which were matched with their corresponding non-neoplastic liver parenchyma (cirrhotic tissues) and 5 normal liver donors using transcriptomic data our group previously published (GSE64041) [28]. CEL files were normalized using the Qlucore software (Qlucore AB, Lund, Sweden) [29]. HMGA1 expression was extracted for each sample.
Expression of HMGA1 by Quantitative Real-Time PCR
RNA from 13/37 matched biopsies of HCC and their cirrhotic tissue previously subjected to transcriptomic profiling [28] was available and subjected to qRT-PCR analysis (Supplementary Methods).
Immunohistochemistry
Immunohistochemical staining of HMGA1 was assessed on a TMA of an independent cohort of 192 HCCs, 108 cirrhotic tissues and 79 normal liver samples, as previously described [29], [30]. Follow-up information was available for 100/192 patients with HCC. HMGA1 antibody was raised against a synthetic peptide as previously reported [19]. Staining was performed as described previously [19], [25] (Supplementary Methods). Samples with ≥5% HMGA1-positive cells were considered HMGA1-positive [19], [25]. Staining was independently scored by three pathologists (DB, FT and LT).
Statistical Analysis
Statistical analyses for categorical and non-categorical variables were performed using Chi-Square/Fisher's Exact and Mann–Whitney U/Student's t tests. Analysis of the variance was performed using the ANOVA test. Correlation was assessed using Spearman's rank correlation. Survival analyses were performed using the Kaplan–Meier method and the log rank test. All tests were two-sided. P-values <0.05 were considered statistically significant. All analyses were performed using Graphpad Prism 6.0 (Graphpad Software, Inc., La Jolla, CA) or SPSS v.20 (Endicott, New York, NY).
Cell Lines
Four liver cancer cell lines (PCL5, HEPG2, SNU449 and SNU182) were used for in vitro experiments. All cell lines were negative for mycoplasma infection using the Universal Mycoplasma Detection kit (ATCC, Manassas, VA). Culture conditions are described in Supplementary Methods.
Vector Construction, Transfections of Mammalian Cells and Analysis of Transgene Expression
For overexpression, the pCDNA3.1-HMGA1 and the empty control vectors were constructed as previously described [10]. For down-regulation, the hairpin RNA interference plasmid for human HMGA1 (pLKO.1-HMGA1, TRCN0000018949) and the scramble control pLKO.1-Puro plasmid (SHC002) were purchased from Sigma-Aldrich (Sigma-Aldrich, St. Louis, MO). The expression of HMGA1 in stable clones was evaluated by western blot (Supplementary Methods).
Proliferation Assay
Proliferation assays were performed using the xCELLigence Real-Time Cell Analysis (RTCA, ACEA Biosciences, San Diego, CA, USA) system. Cell index values were calculated and normalized by the RTCA Software Package v.1.2 (Supplementary Methods). Numerical data were expressed as mean ± standard deviation. Growth curves were analyzed using multiple t-tests, corrected for multiple comparisons by the Holm-Šídák method (alpha: 0.05) using GraphPad Prism 6.0 (Graphpad Software, Inc.).
Transwell Migration Assay
The transwell migration assay was used to assess the chemotactic and migration capacity of cells. Cells were seeded in the upper part of the transwell (8 μm pore membranes) of the 24-well plate and higher serum content was placed in the lower compartment to attract cells to migrate through the membrane. Cells that passed through the membrane were fixed on the membrane using methanol (Supplementary Methods). Fixed cells were stained with crystal violet and the number of migrated cells was determined by the Benchmark Plus microplate spectrophotometer (Bio-Rad, Hercules, CA, USA).
Soft Agar Colony Formation Assay
To assess cellular anchorage-independent growth in vitro, the soft agar colony formation assay, a stringent method for the detection of the tumorigenic potential was used for the pCDNA3.1-HMGA1 and the pLKO.1-HMGA1 transformed cells (Supplementary Methods). Statistical analyses of the number and size of the colonies were performed with GraphPad Prism 6.0 (Graphpad Software, Inc.) using the Student's t test with Welch correction.
Results
HMGA1 mRNA is Frequently Up-Regulated in HCC
To determine the expression level of HMGA1 in HCC, we re-analyzed a published gene expression microarray dataset of 59 HCC biopsies, 37 of which were matched with their respective non-tumoral cirrhotic tissues and 5 normal liver donor samples (GSE64041) [28]. Notably, none of the patients involved in the study received any therapeutic anti-cancer treatment at the time of biopsy.
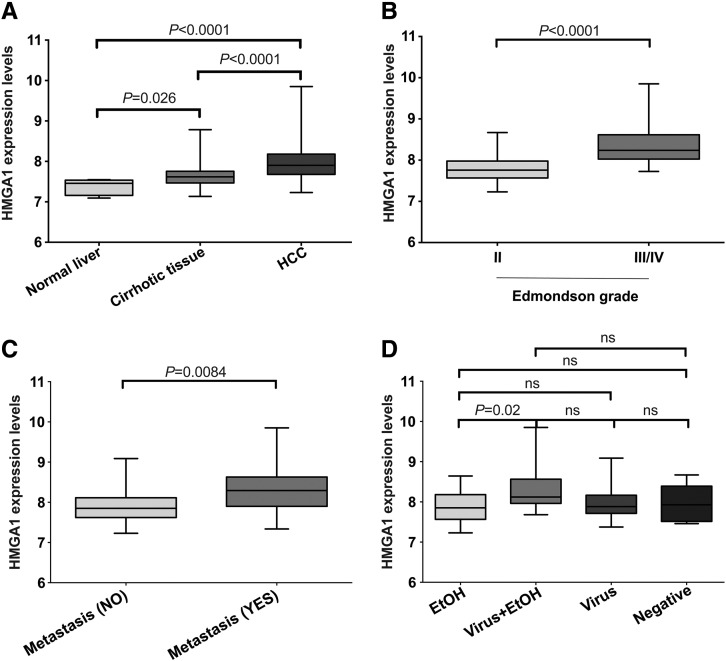
Our analysis showed a monotonic increase in HMGA1 mRNA expression from normal liver tissues to cirrhotic tissues to HCCs (P < .001; ANOVA test; Figure 1A), with increased expression in HCCs compared to cirrhotic tissues and normal liver tissues (both P < .0001, Mann-Whitney U tests) and in cirrhotic tissues compared to normal liver tissues (P = .026, Mann-Whitney U test; Figure 1A). Moreover, HMGA1 expression levels were higher in Edmondson grades III/IV than in grade II tumors (P < .0001, Mann-Whitney U test; Figure 1B) and in HCCs with metastasis (regional lymph node invasion and/or distant organ involvement) than those without (P = .0084, Mann-Whitney U test; Figure 1C). When stratified by underlying virus or alcohol background, we found no difference in HMGA1 expression levels, except that HCCs on a combination of virus and alcohol background showed higher expression than HCCs on an alcohol background (P = .02, Mann-Whitney U test; Figure 1D).
Figure 1.
HMGA1 mRNA expression level using gene expression microarrays. Boxplots show HMGA1 expression (A) in HCC area, corresponding non-tumoral area (cirrhotic tissue) and normal liver samples, (B) in moderately and poorly differentiated HCCs, (C) in HCCs associated with and not associated with metastasis and (D) in HCCs stratified according to the etiology. Statistical comparisons were performed using Mann-Whitney U tests. P < .05 was considered statistically significant. EtOH: alcohol-related. ns: not significant.
To confirm the array-derived data, we evaluated the HMGA1 mRNA expression levels in 13 of the 37 paired HCCs and cirrhotic tissues for which RNA was available. Consistent with the array-derived data, we found that HMGA1 expression was higher in HCCs than in cirrhotic tissues (P = .0119; paired Mann-Whitney U test; Supplementary Figure 1A). Furthermore, HMGA1 expression levels from array-derived and qRT-PCR data were highly correlated (r = 0.67, Spearman correlation, Supplementary Figure 1B). Altogether, these results demonstrate that HMGA1 mRNA is overexpressed in HCCs than their matched cirrhotic tissues and normal liver tissues.
HMGA1 Protein Expression is Associated With Disease Progression and Poorer Survival in HCC
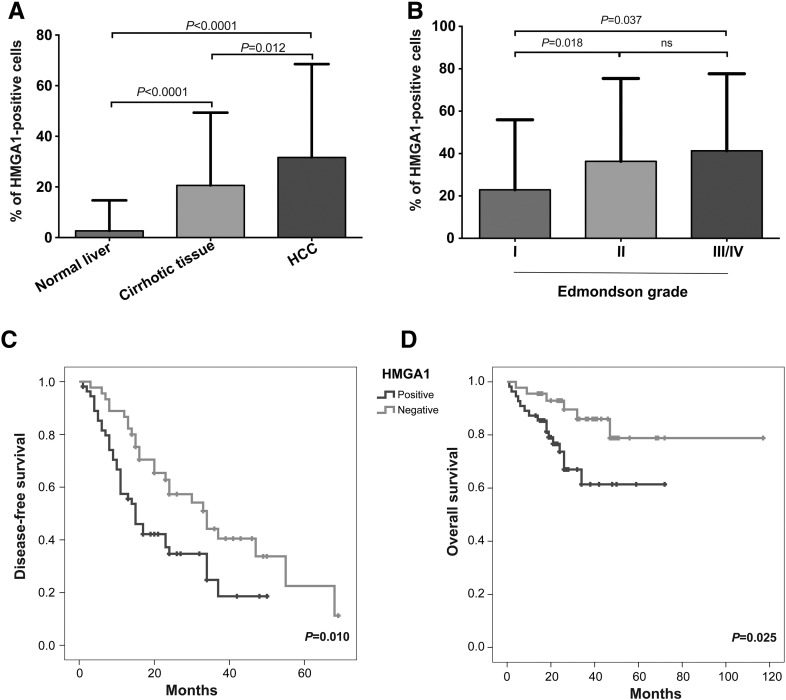
To corroborate our mRNA expression-derived results on the protein level, we evaluated HMGA1 on a TMA of an independent cohort of 192 HCCs, 108 cirrhotic tissues and 79 normal liver samples [29], [30]. Consistent with the mRNA expression analysis, the percentage of HMGA1-positive cells increased monotonically through the progression stages from normal liver to cirrhotic tissue to HCC (P < .0001; ANOVA test; Figure 2A) and were significantly higher in HCCs and in cirrhotic tissues than in normal liver samples (both P < .0001, Mann–Whitney U tests; Figs. 2a and 3). HMGA1 expression was also increased in Edmondson grades III/IV and II HCCs compared to grade I HCCs (P = .037 and P = .018, respectively, Mann–Whitney U tests; Figure 2B) with a monotonic increase from grade I to grades III/IV (P = .026; ANOVA test). Overall, 51%, 42% and 5% of HCCs, cirrhotic tissues and normal liver samples expressed HMGA1, respectively (P < .0001, Chi-squared test; Table 1) and 74%, 52% and 43% of Edmonson grades III/IV, II and I HCCs expressed HMGA1, respectively (P = .031, chi-squared test; Table 2). There was no association between HMGA1 positivity and other clinicopathologic parameters, except the male gender (Table 2).
Figure 2.
HMGA1 protein expression level using TMA. Boxplots show HMGA1 expression (A) in HCCs, cirrhotic tissues and normal liver samples, and (B) in well, moderately and poorly differentiated HCCs. (C) Disease-free and (D) overall survival of patients with HCCs that expressed and did not express HMGA1 using the Kaplan-Meier method. Statistical comparisons were performed using (A and B) Mann-Whitney U tests and (C and D) log-rank tests. P < .05 was considered statistically significant. ns: not significant.
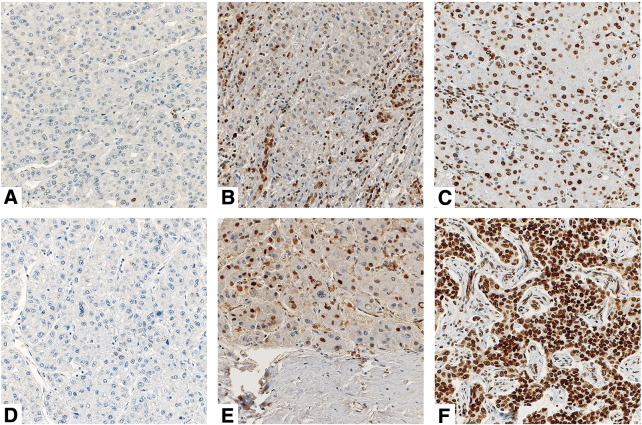
Figure 3.
Representative micrographs of HMGA1 protein staining. Representative micrographs of (A) negative, (B) moderate and (C) high HMGA1 expression in cirrhotic tissues, and of (D) negative, (E) moderate and (F) high HMGA1 expression in HCC. All the micrographs were taken at 20×.
Table 1.
Analysis of HMGA1 Expression by Immunohistochemistry
| HMGA1-negative | HMGA1-positive | % HMGA1-positive | P-value | |
|---|---|---|---|---|
| Normal Liver (n = 79) | 75 | 4 | 5% | <.0001 |
| Cirrhotic tissue (n = 108) | 63 | 45 | 42% | |
| HCC (n = 192) | 95 | 97 | 51% |
Statistical Comparison was Performed Using Chi-Squared Test
Table 2.
Analysis of HMGA1 Expression by Immunohistochemistry in 192 HCCs
| Clinicopathologic information | HMGA1-negative | HMGA1-positive | % HMGA1-positive | P-value | |
|---|---|---|---|---|---|
| Gender | Female | 27 | 14 | 34% | .021 |
| Male | 67 | 84 | 56% | ||
| Tumor Stage | I/II | 58 | 63 | 52% | .869 |
| III/IV | 24 | 28 | 54% | ||
| N stage | 0 | 83 | 88 | 51% | .378 |
| 1 | 4 | 8 | 67% | ||
| M stage | 0 | 71 | 85 | 54% | .214 |
| 1 | 16 | 11 | 41% | ||
| Multifocality | No | 41 | 43 | 51% | 1 |
| Yes | 49 | 52 | 51% | ||
| Vascular Invasion | No | 53 | 53 | 50% | .526 |
| Yes | 28 | 35 | 56% | ||
| Etiology | EtOH | 17 | 19 | 53% | .788 |
| Virus | 43 | 54 | 56% | ||
| Other | 4 | 3 | 43% | ||
| Edmondson grade | I | 44 | 33 | 43% | .031 |
| II | 42 | 45 | 52% | ||
| III/IV | 6 | 17 | 74% | ||
Statistical Comparisons were Performed Using Chi-Squared Tests
We further explored whether HMGA1 positivity was associated with clinical progression and outcome in HCCs. Of the 192 HCCs, clinical follow-up was available for 100 patients. We found that patients with HMGA1-positive HCCs were associated with earlier disease progression and worse overall survival (P = .01 and P = .025, respectively, log rank tests, Figure 2, C and D).
HMGA1 Overexpression Promote Tumor Growth and Migration in In Vitro Models
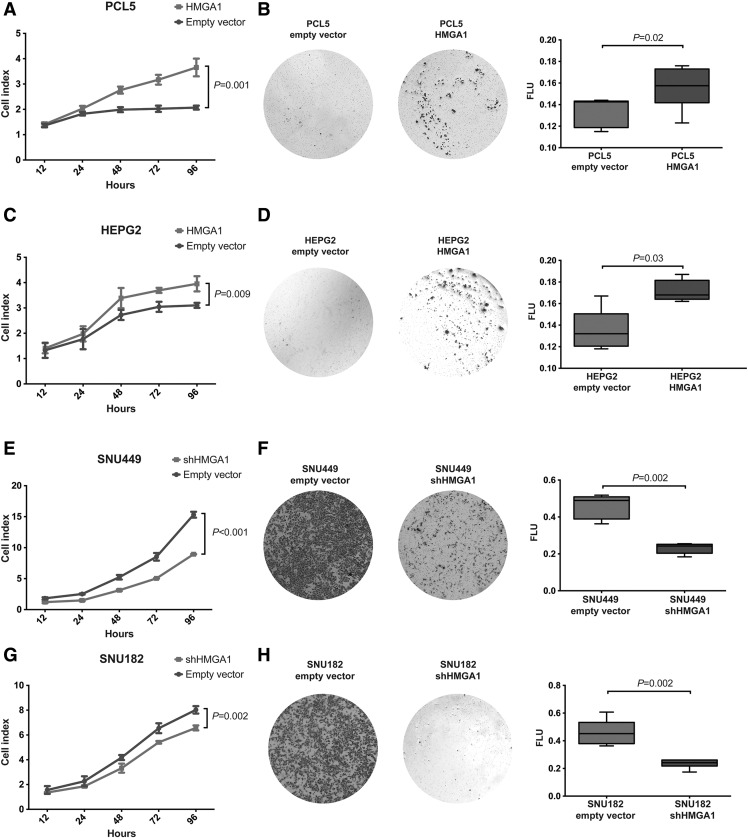
To define whether HMGA1 would have oncogenic properties in in vitro models of HCC, we tested the effect of its overexpression using cDNA constructs in stable cultures of PCL5 and HEPG2 with low HMGA1 endogenous expression and of its silencing in stable cultures of SNU449 and SNU182 with high HMGA1 endogenous expression (Supplementary Figure 2). Forced expression of HMGA1 in PCL5 and HEPG2 led to increased cell growth (both P < .01, Figure 4, A and C), while HMGA1 silencing in SNU449 and SNU182 showed reduced cell growth (both P < .01, Figure 4, E and G) suggesting that HMGA1 expression promotes liver cancer cell growth. Using a transwell migration assay, forced expression of HMGA1 in PCL5 and HEPG2 resulted in increased cell migration (both P < .05, Figure 4, B and D) and conversely, HMGA1 silencing in SNU449 and SNU182 led to decreased cell migration (both P < .01, Figure 4, F and H).
Figure 4.
Impact of HMGA1 on cell growth and migration in in vitro models. Effect of overexpression of HMGA1 in (A) PCL5 and (C) HEPG2 and down-regulation of HMGA1 in (E) SNU449 and (G) SNU182 on cell growth compared to cells transfected with empty vector control. Effect of overexpression of HMGA1 in (B) PCL5 and (D) HEPG2 and down-regulation of HMGA1 in (F) SNU449 and (h) SNU182 on cell migration using Transwell assays compared to cells transfected with empty vector. Quantification was performed using a spectrophotometer (FLU: Fluorescence spectroscopy of dyes). All experiments were performed in triplicates. Error bars, SD of the mean. Statistical comparisons were performed using (A, C, E, G) Holm-Šídák-corrected multiple t tests and (B, D, F, H) Mann-Whitney U tests. P < .05 was considered statistically significant.
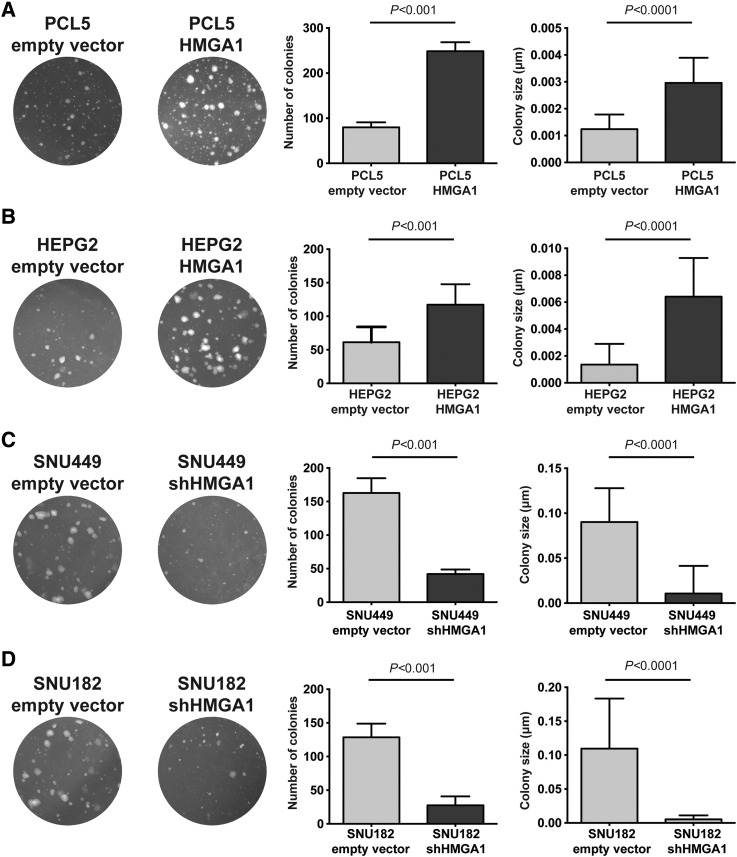
We next investigated whether modulating HMGA1 expression would alter the transformation capacity of liver cancer cells. By assessing the impact of HMGA1 on anchorage-independent growth on soft agar, we found that PCL5 and HEPG2 cells stably overexpressing HMGA1 led to increased number and size of colonies (both at P < .001, Student t-tests with Welch correction; Figure 5, A and B). By contrast, down-regulating HMGA1 in stable clones of SNU449 and SNU182 led to decreased number and size of colonies (both at P < .001, Student t tests with Welch correction; Figure 5, C and D).
Figure 5.
Impact of HMGA1 on cell transformation in in vitro models. Effect of overexpression of HMGA1 in (A) PCL5 and (B) HEPG2 and down-regulation of HMGA1 in (C) SNU449 and (D) SNU182 on anchorage-independent growth. Quantification was performed by defining the number and size of colonies. All experiments were performed in triplicates. Error bars, SD of the mean. Statistical comparisons were performed using unpaired t-tests with Welch correction. P < .05 was considered statistically significant.
Taken together our results suggest that, in line with previous reports in other cancer entities, HMGA1 overexpression plays a pivotal role in cell viability enhancing cell growth and migration and induces transformation by anchorage-independent growth in liver cancer cell lines.
Discussion
In this study, we demonstrated in two independent cohorts that HMGA1 levels monotonically increased through the stages of progression from normal liver to cirrhosis to HCC, both at the mRNA and the protein levels and that HMGA1 protein expression was associated with poor disease-free and overall survival. Our hypothesis that HMGA1 is a driver of progression of HCC is supported by our functional evidence that HMGA1 promoted cell growth, migration and transformation in liver cancer cell lines. These results provide evidence that HMGA1 confers a neoplastic advantage to liver cancer cell lines.
Of particular interest is that HMGA1 is expressed in 42% of cirrhotic tissues. HMGA1 expression has been observed in other preneoplastic conditions, including colon adenomas, pancreatic intraepithelial neoplasias and breast hyperplasia [16], [18], [19]. In fact, a similar pattern of monotonically increasing HMGA1 expression through progression has been found in normal colon epithelium, colon adenoma and colorectal carcinoma [16] and in normal pancreatic tissue, pancreatic intraepithelial neoplasias and invasive ductal adenocarcinoma of the pancreas [19]. Crucially, the substantial proportion of cirrhotic tissues showing high HMGA1 expression suggests that although HMGA1 is a driver of progression, it is not a specific biomarker for HCC.
Consistent with the reported survival differences between HCCs that did or did not express HMGA1 mRNA [27], we found that 51% of HCCs were HMGA1-positive by IHC and that HMGA1 protein expression confers worse prognosis in HCC. While the previous study reported that 13% of HCCs were HMGA1-positive by IHC [27], the higher frequency of HMGA1 positivity we found may be attributed to a much larger cohort and a different antibody for immunodetection. In terms of prognosis, we found that HMGA1 expression conferred worse prognosis, similar to the association of shorter survival in patients with pancreatic ductal carcinoma that showed strong immunoreactivity [20], and to the association of shorter disease-free and overall survival, as well as an increased risk of distant metastases, in patients with HMGA1-overexpressing uveal melanoma [31]. Here we demonstrated that HMGA1 is up-regulated in a substantial proportion of HCC and its expression is associated with poor prognosis.
Previous studies showed that HMGA1 triggers oncogenic transformation in cultured cells [32] and is associated with aggressive cancer subtypes in animal models in several tumor types [20], [33], [34], [35]. For example, in breast cancer cells, HMGA1 overexpression directly activates genes involved in tumor cell migration and invasion [6] and induces epithelial-to-mesenchymal transition [36]. Indeed, we demonstrated in multiple liver cancer cell line models that the HMGA1 increases tumor cell growth and migration and that HMGA1 increased transformation potential in liver cancer cells. Nonetheless, these results are consistent with our and others' [27] observation that HMGA1 expression was associated with Edmondson grade and support the role of HMGA1 as a driver of progression.
This study has limitations. We studied HMGA1 protein expression on TMA punches rather than whole sections, thus the observed expression may not be representative of the individual tissue samples. Despite this, given the large cohort, we expect that the results to be representative on the cohort level. Secondly, the HCCs in the TMA cohort were from resected materials rather than untreated biopsies, thus the HMGA1 levels may have been altered as a result of the surgical procedures and the long post-surgical hypoxia derived for the lack of blood perfusion. However, the analysis of the expression microarrays of untreated liver biopsies is in agreement with the findings at the protein level and it should be emphasized that the liver biopsies, unlike resected materials, had never been subjected to HCC-tailored therapies and are thus the most representative of the natural biology of HCC. In conclusion, our findings demonstrated a functional role for HMGA1 in the progression of HCC. Given multi-faceted functions of HMGA1, further characterization of its function in liver biology will provide novel insights into its mechanisms of action in driving disease progression.
Conflict of Interest
The authors have no conflicts of interest to declare.
Role of the Funding Source
Funding bodies had no role in the design of the study, collection, analysis and interpretation of the data or the writing of the manuscript.
Footnotes
Funding: The study was supported by grants from Oncosuisse (KLS-3639-02-2015) and the Swiss Cancer Research foundation (KFS-3302-08-2013).
Supplementary data to this article can be found online at doi:10.1016/j.neo.2016.10.002.
Contributor Information
Luigi M Terracciano, Email: Luigi.Terracciano@usb.ch.
Salvatore Piscuoglio, Email: Salvatore.Piscuoglio@usb.ch.
Appendix A. Supplementary data
Supplementary methods
Supplementary Figure 1 (A) HMGA1 expression is increased in HCC area compared to cirrhotic tissues by mean of quantitative real-time PCR. Statistical comparisons performed using paired Mann–Whitney U test. P < .05 was considered statistically significant. (B) Spearman correlation analysis performed by using array-derived and qRT-PCR expression data.
Supplementary Figure 2. (A) HMGA1 expression in liver cancer derived cell lines. Quantitative measurement of HMGA1 using western blot analysis after transfection with pCDNA3.1-HMGA1 (B, C) and pLKO.1-HMGA1 (D).
References
- 1.Bianchi ME, Beltrame M. Upwardly mobile proteins. Workshop: the role of HMG proteins in chromatin structure, gene expression and neoplasia. EMBO Rep. 2000;1(2):109–114. doi: 10.1093/embo-reports/kvd030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reeves R. Structure and function of the HMGI(Y) family of architectural transcription factors. Environ Health Perspect. 2000;108(Suppl. 5):803–809. doi: 10.1289/ehp.00108s5803. [DOI] [PubMed] [Google Scholar]
- 3.Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277(1–2):63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- 4.Thomas JO. HMG1 and 2: architectural DNA-binding proteins. Biochem Soc Trans. 2001;29(Pt 4):395–401. doi: 10.1042/bst0290395. [DOI] [PubMed] [Google Scholar]
- 5.Reeves R, Adair JE. Role of high mobility group (HMG) chromatin proteins in DNA repair. DNA Repair (Amst) 2005;4(8):926–938. doi: 10.1016/j.dnarep.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007;7(12):899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- 7.Fedele M, Fusco A. HMGA and cancer. Biochim Biophys Acta. 2010;1799(1–2):48–54. doi: 10.1016/j.bbagrm.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Zhou G, Chen J, Lee S, Clark T, Rowley JD, Wang SM. The pattern of gene expression in human CD34(+) stem/progenitor cells. Proc Natl Acad Sci U S A. 2001;98(24):13966–13971. doi: 10.1073/pnas.241526198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giancotti V, Berlingieri MT, DiFiore PP, Fusco A, Vecchio G, Crane-Robinson C. Changes in nuclear proteins on transformation of rat epithelial thyroid cells by a murine sarcoma retrovirus. Cancer Res. 1985;45(12 Pt 1):6051–6057. [PubMed] [Google Scholar]
- 10.Puca F, Colamaio M, Federico A, Gemei M, Tosti N, Bastos AU. HMGA1 silencing restores normal stem cell characteristics in colon cancer stem cells by increasing p53 levels. Oncotarget. 2014;5(10):3234–3245. doi: 10.18632/oncotarget.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah SN, Kerr C, Cope L, Zambidis E, Liu C, Hillion J. HMGA1 reprograms somatic cells into pluripotent stem cells by inducing stem cell transcriptional networks. PLoS One. 2012;7(11):e48533. doi: 10.1371/journal.pone.0048533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hillion J, Wood LJ, Mukherjee M, Bhattacharya R, Di Cello F, Kowalski J. Upregulation of MMP-2 by HMGA1 promotes transformation in undifferentiated, large-cell lung cancer. Mol Cancer Res. 2009;7(11):1803–1812. doi: 10.1158/1541-7786.MCR-08-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Resar LM. The high mobility group A1 gene: transforming inflammatory signals into cancer? Cancer Res. 2010;70(2):436–439. doi: 10.1158/0008-5472.CAN-09-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takaha N, Resar LM, Vindivich D, Coffey DS. High mobility group protein HMGI(Y) enhances tumor cell growth, invasion, and matrix metalloproteinase-2 expression in prostate cancer cells. Prostate. 2004;60(2):160–167. doi: 10.1002/pros.20049. [DOI] [PubMed] [Google Scholar]
- 15.Tesfaye A, Di Cello F, Hillion J, Ronnett BM, Elbahloul O, Ashfaq R. The high-mobility group A1 gene up-regulates cyclooxygenase 2 expression in uterine tumorigenesis. Cancer Res. 2007;67(9):3998–4004. doi: 10.1158/0008-5472.CAN-05-1684. [DOI] [PubMed] [Google Scholar]
- 16.Abe N, Watanabe T, Sugiyama M, Uchimura H, Chiappetta G, Fusco A. Determination of high mobility group I(Y) expression level in colorectal neoplasias: a potential diagnostic marker. Cancer Res. 1999;59(6):1169–1174. [PubMed] [Google Scholar]
- 17.Chiappetta G, Manfioletti G, Pentimalli F, Abe N, Di Bonito M, Vento MT. High mobility group HMGI(Y) protein expression in human colorectal hyperplastic and neoplastic diseases. Int J Cancer. 2001;91(2):147–151. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1033>3.3.co;2-m. [DOI] [PubMed] [Google Scholar]
- 18.Chiappetta G, Botti G, Monaco M, Pasquinelli R, Pentimalli F, Di Bonito M. HMGA1 protein overexpression in human breast carcinomas: correlation with ErbB2 expression. Clin Cancer Res. 2004;10(22):7637–7644. doi: 10.1158/1078-0432.CCR-04-0291. [DOI] [PubMed] [Google Scholar]
- 19.Piscuoglio S, Zlobec I, Pallante P, Sepe R, Esposito F, Zimmermann A. HMGA1 and HMGA2 protein expression correlates with advanced tumour grade and lymph node metastasis in pancreatic adenocarcinoma. Histopathology. 2012;60(3):397–404. doi: 10.1111/j.1365-2559.2011.04121.x. [DOI] [PubMed] [Google Scholar]
- 20.Hristov AC, Cope L, Di Cello F, Reyes MD, Singh M, Hillion JA. HMGA1 correlates with advanced tumor grade and decreased survival in pancreatic ductal adenocarcinoma. Mod Pathol. 2010;23(1):98–104. doi: 10.1038/modpathol.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masciullo V, Baldassarre G, Pentimalli F, Berlingieri MT, Boccia A, Chiappetta G. HMGA1 protein over-expression is a frequent feature of epithelial ovarian carcinomas. Carcinogenesis. 2003;24(7):1191–1198. doi: 10.1093/carcin/bgg075. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Z, Wang Q, Chen F, Liu J. Elevated expression of HMGA1 correlates with the malignant status and prognosis of non-small cell lung cancer. Tumour Biol. 2015;36(2):1213–1219. doi: 10.1007/s13277-014-2749-4. [DOI] [PubMed] [Google Scholar]
- 23.Kettunen E, Anttila S, Seppanen JK, Karjalainen A, Edgren H, Lindstrom I. Differentially expressed genes in nonsmall cell lung cancer: expression profiling of cancer-related genes in squamous cell lung cancer. Cancer Genet Cytogenet. 2004;149(2):98–106. doi: 10.1016/S0165-4608(03)00300-5. [DOI] [PubMed] [Google Scholar]
- 24.Franco R, Esposito F, Fedele M, Liguori G, Pierantoni GM, Botti G. Detection of high-mobility group proteins A1 and A2 represents a valid diagnostic marker in post-pubertal testicular germ cell tumours. J Pathol. 2008;214(1):58–64. doi: 10.1002/path.2249. [DOI] [PubMed] [Google Scholar]
- 25.Sepe R, Piscuoglio S, Quintavalle C, Perrina V, Quagliata L, Formisano U. HMGA1 overexpression is associated with a particular subset of human breast carcinomas. J Clin Pathol. 2015 doi: 10.1136/jclinpath-2015-202907. [DOI] [PubMed] [Google Scholar]
- 26.Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44(6):694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang ZG, Yang LY, Wang W, Peng JX, Huang GW, Tao YM. Determination of high mobility group A1 (HMGA1) expression in hepatocellular carcinoma: a potential prognostic marker. Dig Dis Sci. 2005;50(10):1764–1770. doi: 10.1007/s10620-005-2934-9. [DOI] [PubMed] [Google Scholar]
- 28.Makowska Z, Boldanova T, Adametz D, Quagliata L, Vogt JE, Dill MT. Gene expression analysis of biopsy samples reveals critical limitations of transcriptome-based molecular classifications of hepatocellular carcinoma. J Pathol Clin Res. 2016;2:80–92. doi: 10.1002/cjp2.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quagliata L, Andreozzi M, Kovac M, Tornillo L, Makowska Z, Moretti F. SH2D4A is frequently downregulated in hepatocellular carcinoma and cirrhotic nodules. Eur J Cancer. 2014;50(4):731–738. doi: 10.1016/j.ejca.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 30.Baumhoer D, Tornillo L, Stadlmann S, Roncalli M, Diamantis EK, Terracciano LM. Glypican 3 expression in human nonneoplastic, preneoplastic, and neoplastic tissues: a tissue microarray analysis of 4,387 tissue samples. Am J Clin Pathol. 2008;129(6):899–906. doi: 10.1309/HCQWPWD50XHD2DW6. [DOI] [PubMed] [Google Scholar]
- 31.Qu Y, Wang Y, Ma J, Zhang Y, Meng N, Li H. Overexpression of high mobility group A1 protein in human uveal melanomas: implication for prognosis. PLoS One. 2013;8(7):e68724. doi: 10.1371/journal.pone.0068724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wood LJ, Maher JF, Bunton TE, Resar LM. The oncogenic properties of the HMG-I gene family. Cancer Res. 2000;60(15):4256–4261. [PubMed] [Google Scholar]
- 33.Tanaka M, Itoh T, Tanimizu N, Miyajima A. Liver stem/progenitor cells: their characteristics and regulatory mechanisms. J Biochem. 2011;149(3):231–239. doi: 10.1093/jb/mvr001. [DOI] [PubMed] [Google Scholar]
- 34.Sarhadi VK, Wikman H, Salmenkivi K, Kuosma E, Sioris T, Salo J. Increased expression of high mobility group A proteins in lung cancer. J Pathol. 2006;209(2):206–212. doi: 10.1002/path.1960. [DOI] [PubMed] [Google Scholar]
- 35.Flohr AM, Rogalla P, Bonk U, Puettmann B, Buerger H, Gohla G. High mobility group protein HMGA1 expression in breast cancer reveals a positive correlation with tumour grade. Histol Histopathol. 2003;18(4):999–1004. doi: 10.14670/HH-18.999. [DOI] [PubMed] [Google Scholar]
- 36.Dolde CE, Mukherjee M, Cho C, Resar LM. HMG-I/Y in human breast cancer cell lines. Breast Cancer Res Treat. 2002;71(3):181–191. doi: 10.1023/a:1014444114804. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary methods
Supplementary Figure 1 (A) HMGA1 expression is increased in HCC area compared to cirrhotic tissues by mean of quantitative real-time PCR. Statistical comparisons performed using paired Mann–Whitney U test. P < .05 was considered statistically significant. (B) Spearman correlation analysis performed by using array-derived and qRT-PCR expression data.
Supplementary Figure 2. (A) HMGA1 expression in liver cancer derived cell lines. Quantitative measurement of HMGA1 using western blot analysis after transfection with pCDNA3.1-HMGA1 (B, C) and pLKO.1-HMGA1 (D).