FIGURE 4.
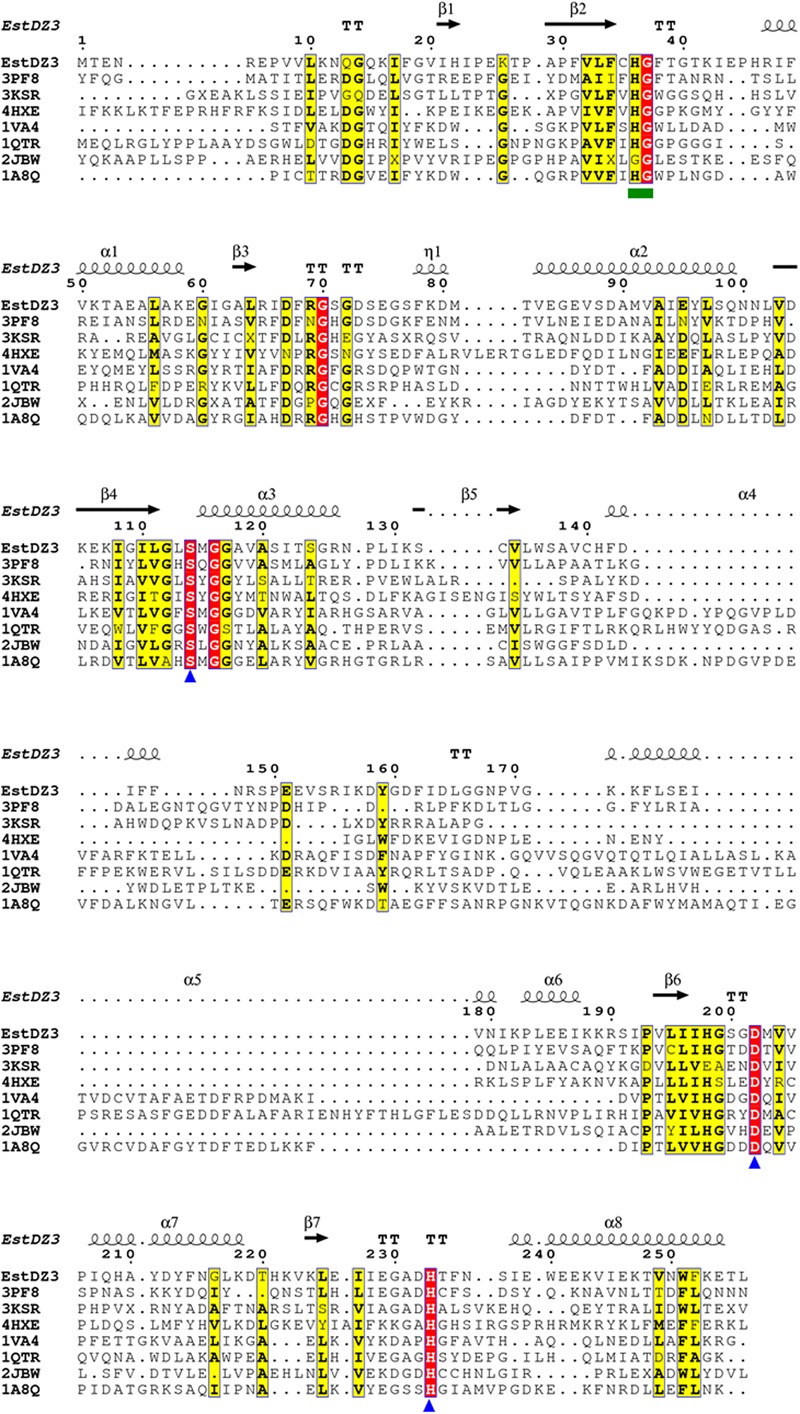
Multiple sequence alignment of EstDZ3 and homologs with known three-dimensional (3D) structure. The absolutely conserved amino acids are highlighted in red and similar ones in yellow. The catalytic residues, Ser114, Asp202, and His233 are indicated by blue triangles. The conserved His36-Gly37 dipeptide, which participates in the formation of the oxyanion hole during ester hydrolysis, is indicated by a green square. Elements of the predicted EstDZ3 secondary structure are denoted as α (α helix), β (β sheet), η (random coil), and T (β turn). Sequence alignment was performed using Clustal Omega (Sievers et al., 2011) and illustrated by ESPript (Robert and Gouet, 2014).
