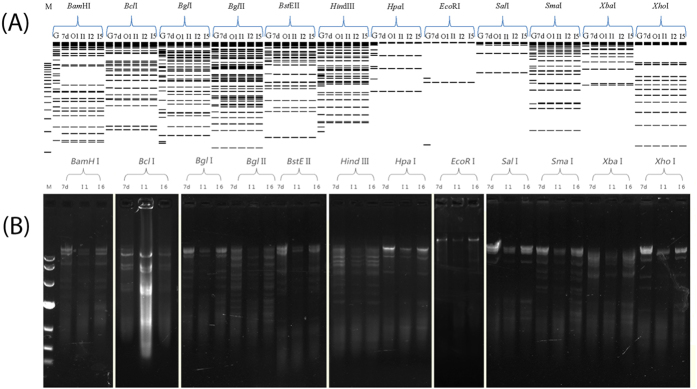
Figure 3. Determination of genome types of HAdV-7 isolates from children with ARD in Guangzhou, Southern China.
The in silico restriction endonuclease analysis (REA) was performed using the Vector NTI 11.5 (Invitrogen Corp; San Diego) to identify the genome types of circulating HAdV-7 isolates OP01_2011 (lane O1; KP670857), IP01_2010 (lane I1; KP670858), IP02_2011 (lane I2; KP670856) and IP05_2012 (lane I5; KP670860) (A). Genome sequences of strains Gomen type 7 prototype Gomen (lane G; AY594255) and DG01_2011 7d (lane 7d; KC440171) were also analyzed with BamHI, BclI, BglI, BglII, BstEII, HindIII, HpaI, EcoRI, SalI, SmaI, XbaI, and XhoI (TaKaRa Corp.; China) respectively, as described earlier (Zhao et al. Sci Rep, 2014, 4; doi:10.1038/srep07365). The agarose gel electrophoresis of RE digested genomic DNA was also performed to confirm the in silico REA (B). IP01_2010 (lane I1; KP670858) and IP06 (lane I6; KP670859) were chosen to be analyzed. All the seven HAdV-7 isolates in this study have identical REA profiles with each other, different from the Gomen prototype, identified as genome type HAdV-7d. M, molecular weight markers corresponding to a 500 bp DNA Ladder (A) and DL10,000 DNA Marker (B) respectively. Nota bene: the agarose gel electrophoresis of RE digested genomic DNA was not run at the same time but under the same conditions.