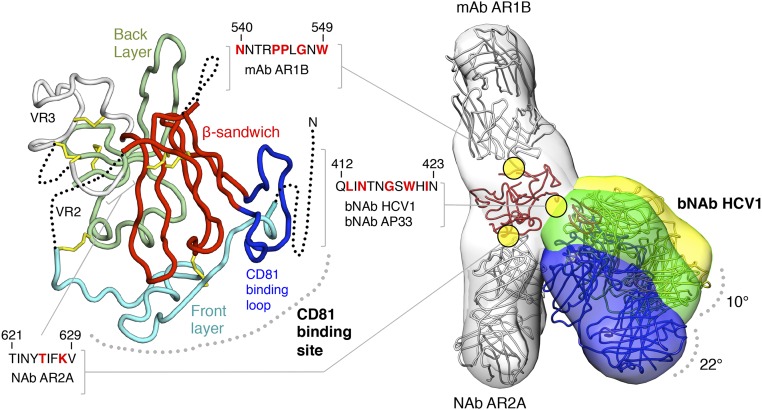
Fig. 1.
Conformational flexibility of a broadly neutralizing epitope on soluble HCV E2c2 revealed by EM. (Left) Crystal structure of E2c is displayed as a ribbon with structural elements indicated and disordered regions shown as dotted lines. The epitopes of mAb AR1B, bNAb HCV1, bNAb AP33, and NAb AR2A are defined; residues that are important for binding, as mapped by alanine scanning experiments, are colored red in the sequences shown. (Right) Three negative-stain EM density maps at ∼30-Å resolution of E2c2 (gray) bound to Fab AR1B (gray), AR2A (gray), and HCV1 (yellow, green, and blue) are shown as transparent surfaces with structural models of the antibodies and E2c superimposed (also Fig. S3). Differences in bNAb HCV1 angles of approach indicated from the three RCT reconstructions are shown.