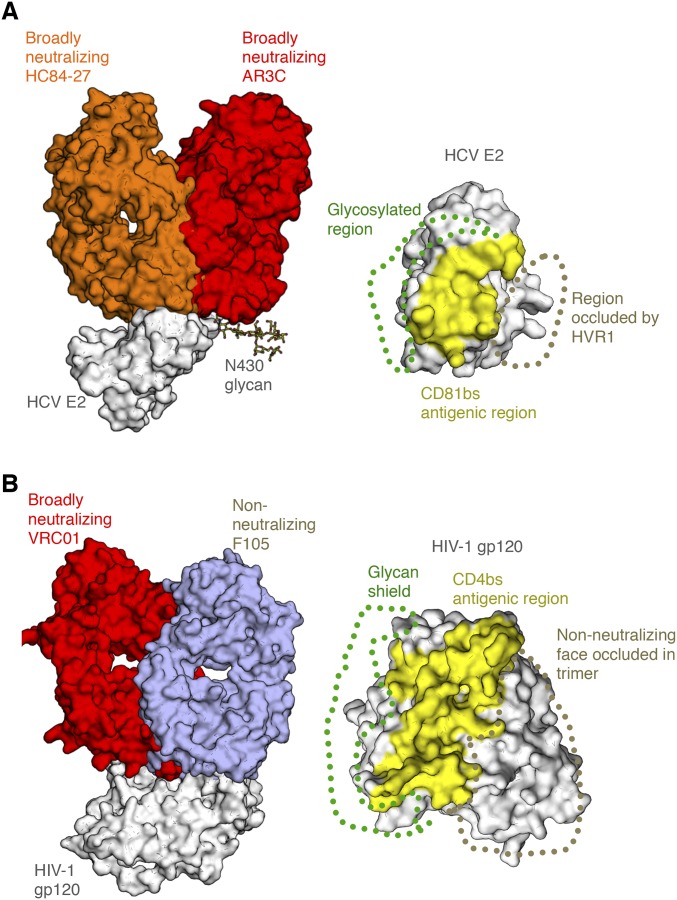
Fig. S1.
Angle of approach by antibodies that recognize the HCV E2 receptor-binding site. (A, Left) Crystal structure bNAb HC84-27 (orange) [Protein Data Bank (PDB) ID code 4JZO] is superposed on its peptide epitope in the crystal structure of the HCV E2 core bound to bNAb AR3C (red) (PDB ID code 4MWF) (gray). (A, Right) CD81bs, targeted by both bNAbs, is highlighted, with the general location of the glycosylated region (glycan shield) and HVR1 indicated. (B) Comparison of HIV Env gp120 epitopes at or around the CD4 receptor-binding site, indicating regions protected by the glycan shield and the Env trimer interface. Crystal structures of bNAb VRC01 (red) (PDB ID code 4LST) and non-NAb F105 (wheat) (PDB ID code 3HI1) are shown superposed on the gp120 core antigen (gray). (Right) Common surface recognized by many bnAbs, the CD4-binding site (CD4bs), is highlighted, with the glycan shield shown, as well as the region occluded within the trimeric spike that is not accessible to non-NAbs in the trimer context.