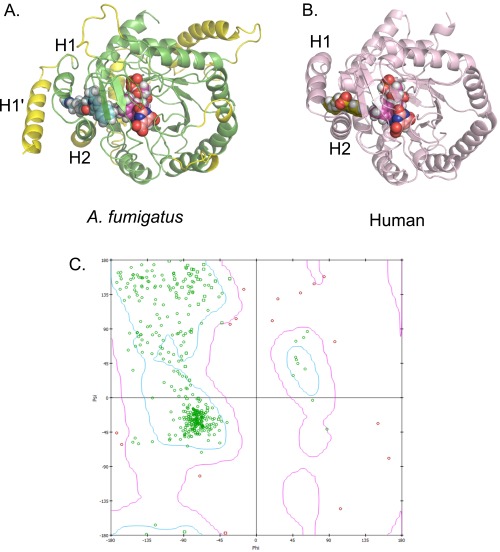
Fig. S6.
Inhibitor binding to DHODH. (A) Homology model of A. fumigatus DHODH with the estimated binding mode of F901318. The A. fumigatus DHODH was modeled using information from known class 2 DHODH structures, with the human structure (PDB ID code 1D3G) forming the protein template. Manually modeled loop/helix insertions are shown in yellow and include an extra N-terminal alpha-helix (H1′) compared with the human protein. Helices H1 and H2 are common to these structures and are predicted to be the entry point for ubiquinone from the mitochondrial membrane (19). F901318 is shown as cyan spheres, FMN is in magenta, and orotate is in orange. (B) Brequinar bound to human DHODH (PDB ID code 1D3G). Adapted from Liu et al. (19). Brequinar is in green; FMN, in magenta; orotate, in orange. (C) Ramachandran plot of an A. fumigatus DHODH homology model. The model was assessed by calculating the ϕ and ψ torsional angles of the amino acid residues in the model. The bulk of the model appears to be energetically allowable (green residues). All residues with ϕ–ψ angle irregularities are exclusively in regions with lower homology to known structures and thus are more speculative. These discrepancies (pink residues) occur in residues distant from the predicted inhibitor-binding region and should not affect the derived ligand-binding hypothesis.