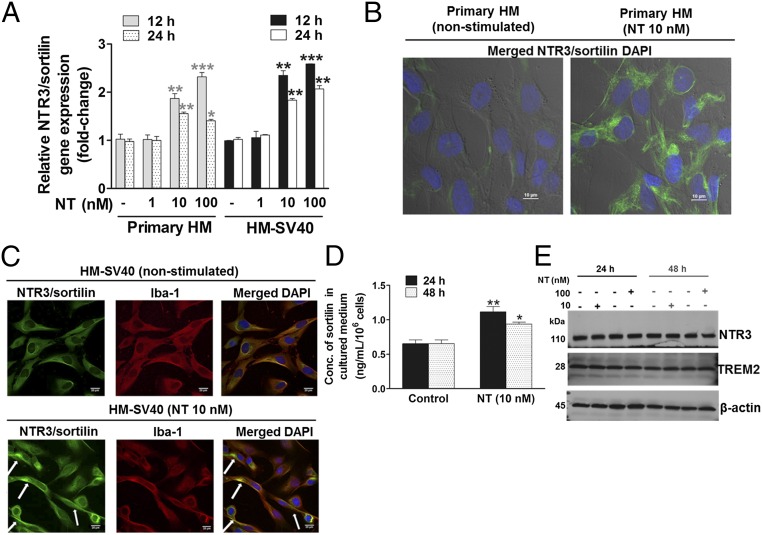
Fig. 3.
NTR3/sortilin gene expression and cellular localization in HM, and its release in response to NT. (A) Primary HM (2.5 × 105 cells) and immortalized HM-SV40 were stimulated with NT for 12 or 24 h to measure gene expression levels of NTR3/sortilin in control and NT-stimulated (10 or 100 nM) microglia by qRT-PCR. All conditions were performed in triplicate for each dataset and were repeated three times (n = 3). Results were normalized against the endogenous GAPDH and expressed relative to the mean of the control for the gene of interest, with significance of comparisons denoted by *P < 0.05, **P < 0.001, or ***P < 0.0001. Microglia-SV40 (5 × 103 cells per four-well chamber–coated slide) were stimulated with NT (10 nM) for 24 h, and then fixed and permeabilized to stain for nuclei using DAPI (blue), with specific antibodies for Alexa 488-NTR3/sortilin (green) or Alexa 594–Iba-1, a microglial marker protein (red), whereas rabbit IgG was used for the negative control. The cell surface and cytosolic distribution of NTR3/sortilin protein is shown in control and NT-stimulated primary HM (B) and immortalized HM-SV40 (C), where colocalization with Iba-1 is also apparent (white arrows). Qualitative analysis was done using images from triplicates, and representative images are shown. (D) HM-SV40 (1 × 106 cells) were stimulated with NT (10 nM) for 24 or 48 h to measure release of soluble NTR3/sortilin in culture media by ELISA. All conditions were performed in triplicate for each dataset and repeated three times (n = 3). Significance of comparisons is denoted by *P < 0.05 or **P < 0.001. (E) HM-SV40 (1 × 106 cells) were stimulated with NT (10 or 100 nM) for 24 or 48 h to measure total cellular NTR3/sortilin levels by Western blot analysis, where TREM-2 served as a microglial marker protein and β-actin as the loading control. All conditions were performed in triplicate for each dataset and were repeated three times (n = 3), and a representative image is shown.