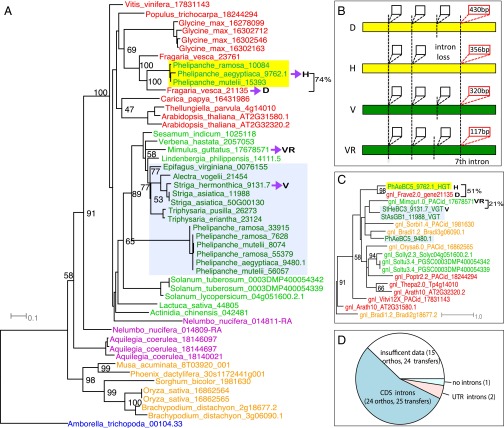
Fig. 3.
Genomic horizontal transfer of a tRNAHis guanylyltransferase from Frave (Fragaria) (or its ancestor) to Phelipanche. (A) A coding-sequence (CDS) tree by RAxML from represented species across angiosperm lineages. D, inferred donor (in Fragaria); H, the parasitic HGT gene; V, vertical parasitic gene; VR, related sequence of the vertical parasitic gene (in Mimulus). Seventy-four percent represents the CDS similarity between the HGT gene and its inferred donor. (B) Gene structure with four selected introns for the four sequences (H, D, V, and VR). Yellow and green bars represent coding sequence; the vertical dashed lines represent the intron positions; the boxes represent introns. At least four conserved intron positions were shown on the gene structure; the third intron was lost in the HGT gene, and the fourth intron on the graph (which is the seventh intron of the Mimulus gene) showed strong sequence similarity between the HGT gene and its donor (marked by red intron boxes with length within). (C) The phylogeny of the seventh intron (marked red in B) from genes on the CDS tree: the HGT gene groups with its donor supported by 98% BS, whereas the vertically inherited gene groups with a close relative (Mimulus sequence). The intron sequence similarity between the HGT (HGT) gene and its donor (D) is 51%, and the intron sequence similarity between the vertical gene (V) and its vertical relative (VR) is 21%. (D) The number of HGT orthogroups that support 25 genomic transfers in 24 HGT orthogroups containing introns in the CDS region (dark blue) and 2 HGT orthogroups containing introns in the UTR region (pink). The remaining orthogroups contain 15 HGT orthogroups (24 transfers) with insufficient genomic data to infer presence of introns (white) and one orthogroup containing HGT gene without introns (light blue).