Figure 1.
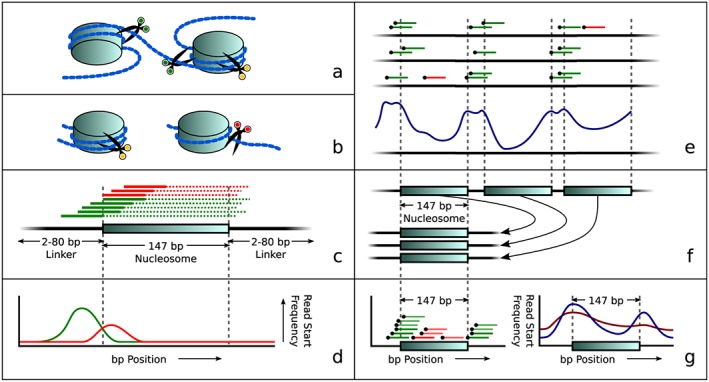
Overview of the principle of our approach: nucleosome‐dependent differences in degradation of maternal and fetal DNA lead to different start sites of sequence reads. (a) Simplified visualization of DNA wound around nucleosomes with three examples of cut positions (green scissors means able to cut in this view while yellow scissors indicate that cutting is protected by the histone complex). (b) Later during degradation, the fragment may either still be protected by a nucleosome (left, yellow scissors), or it may be unwound, allowing it to be cut at the previously inaccessible position (now indicated by red scissors. (c) Theoretical distribution of reads around a nucleosome. Colors correspond to scissor colors in panels (a) and (b). The solid lines depict reads while dotted lines represent the unsequenced parts of these DNA fragments. (d) Idealized read start frequency distribution for reads upstream of the nucleosome center, overstating the difference in positions where cuts were made. (e) Combining several low coverage samples (top) reveals a genome‐wide repetitive pattern (bottom) that matches in size and frequency to nucleosome and linker DNA positions. (f) Aligning all nucleosome positions to a single aligned nucleosome. (g) Read start positions in the aligned nucleosome (left) show the aligned nucleosome profile (right), which can then be used to determine differences in read distributions related to fetal fractions: Blue shows a sample with a low fetal fraction; red shows a sample with a higher fetal fraction
