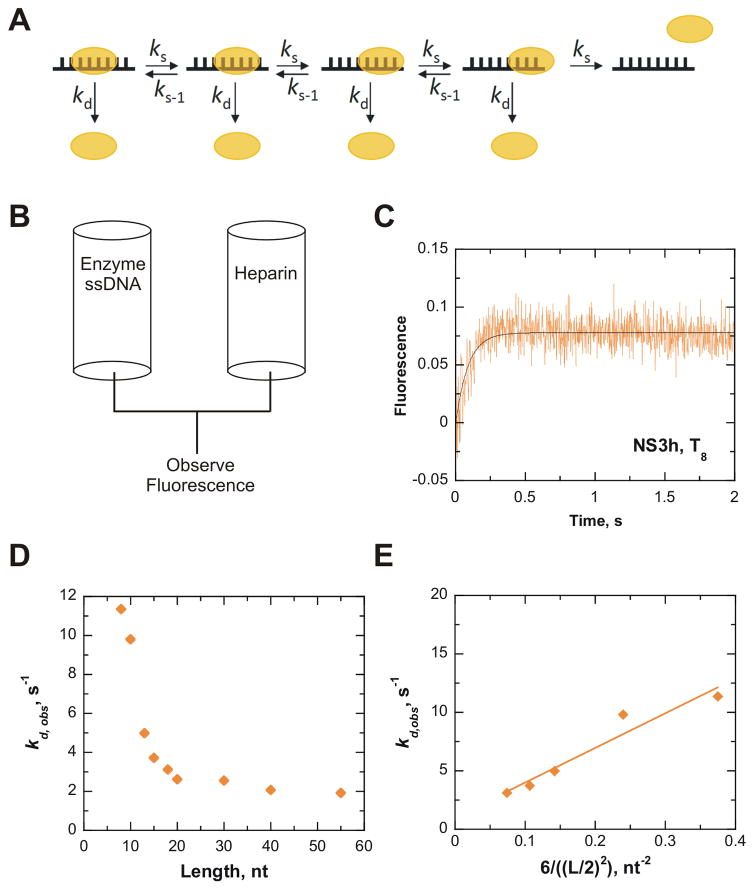
Figure 2.
NS3h slides along ssDNA in an ATP-independent manner. Diffusional sliding of NS3h along ssDNA was determined from dissociation of the enzyme from oligonucleotides of increasing length (L). All concentrations reported are final after mixing. A. A model for diffusion of NS3h along a 8mer oligonucleotide. NS3h binds 6 nt based on the crystal structure. Each step of diffusional sliding (ks) is shown in one direction, but actually can occur with equal probability in both directions. Sliding is assumed to occur in increments of one nt. Dissociation from the end of the DNA was estimated to occur in three individual steps, which allows half the six sites of interaction to dissociate. NS3h can also dissociate from internal positions, kd. B. Protocol for rapid mixing of a solution containing a pre-bound NS3h-ssDNA complex with a solution of heparin, which will sequester any unbound helicase and prevent rebinding of NS3h to the substrate. C. A representative progress curve from dissociation of 200 nM NS3h from 1.2 μM strands T8 upon rapid mixing with 4 mg/mL heparin. The data was fit to a single exponential to obtain an observed dissociation rate (kd,obs) of 11.4 ± 0.8 s−1. D. The kd,obs rates obtained for different lengths of ssDNA are plotted (
 ). E. The rates of dissociation from oligonucleotide that display a length-dependence (T8, T10, T13, T15, T18) were used to determine the rate of sliding. The sliding rate (ks) was determined via analysis of the linear regression fit of the observed dissociation rates plotted versus the equation describing diffusion along a linear polymer.26 The ks of NS3h along ssDNA in the absence of ATP binding and hydrolysis was determined to be 30 nt2/s (
). E. The rates of dissociation from oligonucleotide that display a length-dependence (T8, T10, T13, T15, T18) were used to determine the rate of sliding. The sliding rate (ks) was determined via analysis of the linear regression fit of the observed dissociation rates plotted versus the equation describing diffusion along a linear polymer.26 The ks of NS3h along ssDNA in the absence of ATP binding and hydrolysis was determined to be 30 nt2/s (
 ).
).