Figure 1. Tight Binding of Cas5c to the Pre-crRNA Repeat Is Mediated by the First 4–5 Nucleotides of the 5′ Handle Region.
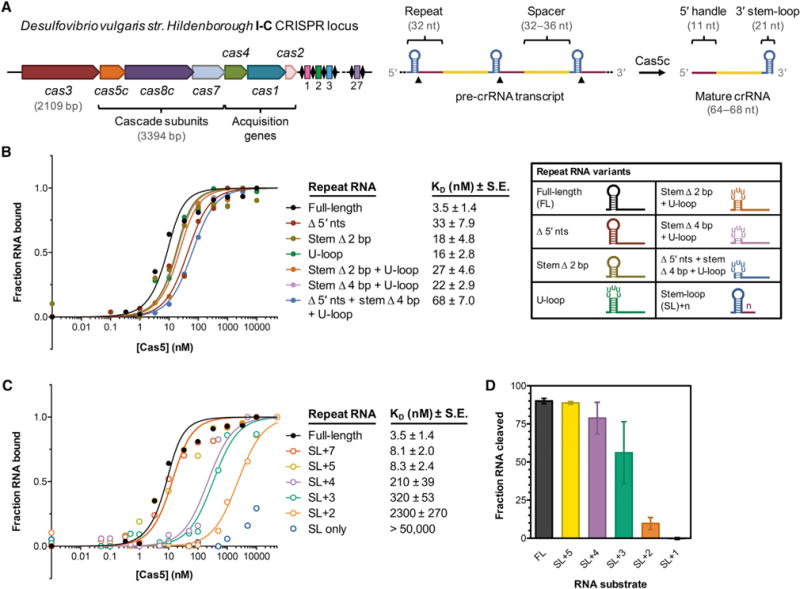
(A) Overview of the type I-C cas genes and CRISPR array found on the Desulfovibrio vulgaris str. Hildenborough megaplasmid. The length of each cas gene is drawn approximately to scale, whereas the repeat-spacer array is drawn much larger for clarity.
(B) At left, normalized florescence polarization (FP) measurements of Cas5c binding to the full-length pre-crRNA repeat and several variants in which the 5′ end and stem-loop have been altered. Cas5c was incubated with each 5′ fluorescein-labeled RNA for 30 min at 37°C prior to FP measurement. Each point represents the average of at least three independent replicates and calculated dissociation constant values (KD) are listed along with the standard error of the fit parameter (SE). Error bars representing ±1 SD are omitted for clarity but can be found in Figure S1C. At right, graphical depictions of the modified repeat sequences tested.
(C) Normalized fluorescence polarization binding data for Cas5c binding to pre-crRNA repeats in which the 5′ handle is progressively truncated. Data represent at least three independent replicates. Error bars representing ±1 SD are omitted for clarity but can be found in Figure S1C.
(D) Quantified cleavage efficiencies of several pre-crRNA variants by Cas5c after incubation for 1 hr at 37°C. Each bar is the average of at least three independent replicates; error bars represent ±1 SD.
See also Figure S1 and Tables S1 and S2.
