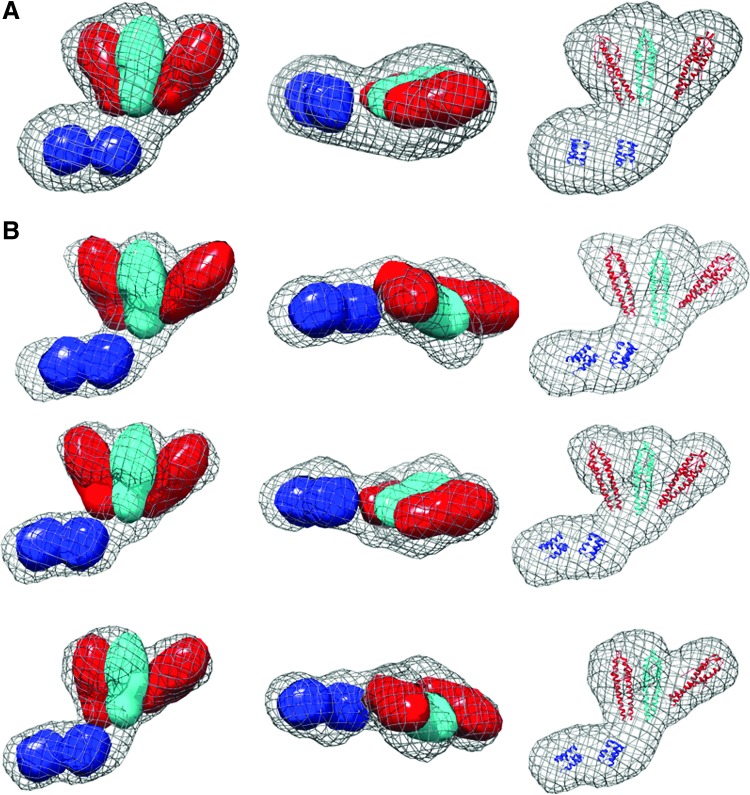
FIG. 3.
SAXS molecular envelopes for LmGpsB. (A) The average surface of 15 independently generated ab initio LmGpsB dummy atom models is represented as a mesh. Three of these models are represented in (B) to illustrate the level of variation. The coordinates of N-LmGpsB dimers (red and cyan) and the C-LmGpsB trimers (blue) were docked manually and are shown in ribbon form (right column) or as equivalent electron density maps would appear at 25 Å resolution (left, middle columns). The coordinates shown correspond to residues 5–67 (N-terminal domain) and 89–106 (C-terminal domain) of LmGpsB; LmGpsB has a total of 113 residues. The models do not include the linker between domains. In the arrangements shown in (B), with the simplest of the possible connectivities between domains, the chain termini are separated by distances of up to 55 Å. By comparison, a fully extended 21 polypeptide residue chain would be ∼80 Å in length, and therefore, the models are compatible with the domains being connected by an appropriate length linker in a semicompact conformation.